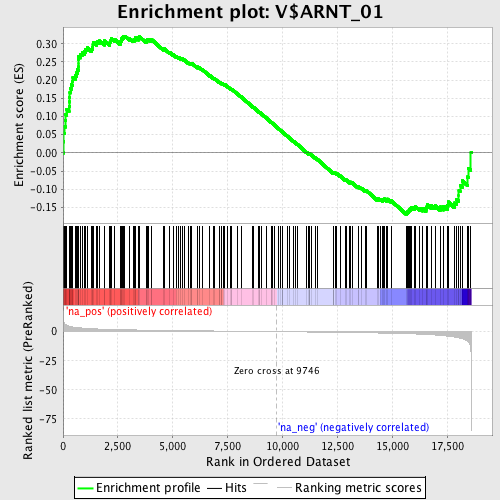
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$ARNT_01 |
| Enrichment Score (ES) | 0.32095134 |
| Normalized Enrichment Score (NES) | 1.4042501 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.18151337 |
| FWER p-Value | 0.986 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SNX8 | 11 | 10.576 | 0.0305 | Yes | ||
| 2 | VARS2 | 25 | 8.561 | 0.0549 | Yes | ||
| 3 | HNRPA1 | 80 | 6.737 | 0.0718 | Yes | ||
| 4 | IFRD2 | 98 | 6.290 | 0.0894 | Yes | ||
| 5 | NUDC | 120 | 5.874 | 0.1055 | Yes | ||
| 6 | PPAT | 158 | 5.554 | 0.1198 | Yes | ||
| 7 | SHMT1 | 272 | 4.709 | 0.1275 | Yes | ||
| 8 | EIF4B | 291 | 4.621 | 0.1401 | Yes | ||
| 9 | AATF | 299 | 4.568 | 0.1532 | Yes | ||
| 10 | QTRT1 | 309 | 4.524 | 0.1660 | Yes | ||
| 11 | RCL1 | 336 | 4.377 | 0.1774 | Yes | ||
| 12 | SLC39A11 | 379 | 4.227 | 0.1876 | Yes | ||
| 13 | NR1D1 | 421 | 4.076 | 0.1973 | Yes | ||
| 14 | HSPD1 | 446 | 3.986 | 0.2077 | Yes | ||
| 15 | DDX3X | 560 | 3.707 | 0.2125 | Yes | ||
| 16 | PSME3 | 617 | 3.558 | 0.2199 | Yes | ||
| 17 | AKAP12 | 650 | 3.499 | 0.2285 | Yes | ||
| 18 | MTCH2 | 692 | 3.421 | 0.2363 | Yes | ||
| 19 | TBC1D5 | 700 | 3.410 | 0.2459 | Yes | ||
| 20 | BOK | 702 | 3.410 | 0.2559 | Yes | ||
| 21 | EIF2C2 | 713 | 3.387 | 0.2653 | Yes | ||
| 22 | PAICS | 798 | 3.244 | 0.2703 | Yes | ||
| 23 | EIF4E | 869 | 3.123 | 0.2757 | Yes | ||
| 24 | AMPD2 | 980 | 2.947 | 0.2784 | Yes | ||
| 25 | CLN3 | 1012 | 2.914 | 0.2852 | Yes | ||
| 26 | SYNCRIP | 1090 | 2.817 | 0.2893 | Yes | ||
| 27 | DSCAM | 1305 | 2.544 | 0.2852 | Yes | ||
| 28 | UBE1 | 1345 | 2.502 | 0.2905 | Yes | ||
| 29 | GATA4 | 1361 | 2.484 | 0.2969 | Yes | ||
| 30 | EIF3S10 | 1365 | 2.478 | 0.3041 | Yes | ||
| 31 | RAB3IL1 | 1514 | 2.334 | 0.3029 | Yes | ||
| 32 | PER1 | 1581 | 2.278 | 0.3060 | Yes | ||
| 33 | VLDLR | 1667 | 2.221 | 0.3079 | Yes | ||
| 34 | CCAR1 | 1883 | 2.075 | 0.3024 | Yes | ||
| 35 | PPRC1 | 1892 | 2.070 | 0.3080 | Yes | ||
| 36 | OPRS1 | 2136 | 1.935 | 0.3005 | Yes | ||
| 37 | HOXB4 | 2137 | 1.934 | 0.3062 | Yes | ||
| 38 | GTF2H1 | 2175 | 1.914 | 0.3098 | Yes | ||
| 39 | ETV1 | 2183 | 1.911 | 0.3151 | Yes | ||
| 40 | SYT3 | 2348 | 1.827 | 0.3115 | Yes | ||
| 41 | UBXD3 | 2601 | 1.714 | 0.3029 | Yes | ||
| 42 | SNX2 | 2625 | 1.698 | 0.3067 | Yes | ||
| 43 | STMN1 | 2649 | 1.690 | 0.3104 | Yes | ||
| 44 | TEF | 2673 | 1.680 | 0.3141 | Yes | ||
| 45 | COPS7A | 2711 | 1.662 | 0.3170 | Yes | ||
| 46 | RHEBL1 | 2751 | 1.643 | 0.3197 | Yes | ||
| 47 | TCERG1 | 2816 | 1.619 | 0.3210 | Yes | ||
| 48 | CD164 | 3024 | 1.535 | 0.3142 | No | ||
| 49 | LTBR | 3198 | 1.474 | 0.3092 | No | ||
| 50 | CHD4 | 3237 | 1.460 | 0.3114 | No | ||
| 51 | CELSR3 | 3283 | 1.444 | 0.3132 | No | ||
| 52 | POGK | 3300 | 1.437 | 0.3166 | No | ||
| 53 | RBBP6 | 3422 | 1.390 | 0.3141 | No | ||
| 54 | COMMD3 | 3444 | 1.382 | 0.3170 | No | ||
| 55 | PPP1R1B | 3479 | 1.372 | 0.3192 | No | ||
| 56 | SYT6 | 3793 | 1.270 | 0.3060 | No | ||
| 57 | UBE4B | 3801 | 1.267 | 0.3093 | No | ||
| 58 | TCEAL1 | 3840 | 1.255 | 0.3109 | No | ||
| 59 | HMGN2 | 3884 | 1.241 | 0.3123 | No | ||
| 60 | PRPS1 | 4009 | 1.203 | 0.3091 | No | ||
| 61 | ATP6V1C1 | 4046 | 1.192 | 0.3106 | No | ||
| 62 | ZCCHC7 | 4584 | 1.059 | 0.2846 | No | ||
| 63 | PPP1R3C | 4605 | 1.055 | 0.2866 | No | ||
| 64 | CNNM1 | 4845 | 0.994 | 0.2766 | No | ||
| 65 | HIRA | 5017 | 0.955 | 0.2701 | No | ||
| 66 | NEUROG2 | 5145 | 0.925 | 0.2660 | No | ||
| 67 | AP1S2 | 5265 | 0.892 | 0.2621 | No | ||
| 68 | ARRDC3 | 5353 | 0.872 | 0.2600 | No | ||
| 69 | ATP6V1A | 5420 | 0.858 | 0.2589 | No | ||
| 70 | RPL13A | 5528 | 0.833 | 0.2556 | No | ||
| 71 | MBNL1 | 5726 | 0.794 | 0.2472 | No | ||
| 72 | NEUROD2 | 5814 | 0.776 | 0.2448 | No | ||
| 73 | SNAP25 | 5828 | 0.775 | 0.2463 | No | ||
| 74 | XPO1 | 5864 | 0.766 | 0.2467 | No | ||
| 75 | ZBTB10 | 6115 | 0.716 | 0.2352 | No | ||
| 76 | PLCG2 | 6130 | 0.712 | 0.2366 | No | ||
| 77 | DNMT3A | 6216 | 0.696 | 0.2340 | No | ||
| 78 | MYST3 | 6354 | 0.664 | 0.2285 | No | ||
| 79 | PICALM | 6681 | 0.598 | 0.2126 | No | ||
| 80 | NKX2-2 | 6855 | 0.561 | 0.2049 | No | ||
| 81 | HOXD10 | 6913 | 0.548 | 0.2034 | No | ||
| 82 | FOXD3 | 7106 | 0.510 | 0.1945 | No | ||
| 83 | PCDHA10 | 7146 | 0.502 | 0.1938 | No | ||
| 84 | PTMA | 7233 | 0.486 | 0.1906 | No | ||
| 85 | CRY2 | 7235 | 0.485 | 0.1920 | No | ||
| 86 | PAK2 | 7318 | 0.467 | 0.1889 | No | ||
| 87 | ATP6V0B | 7374 | 0.457 | 0.1873 | No | ||
| 88 | TBC1D15 | 7493 | 0.434 | 0.1821 | No | ||
| 89 | JPH1 | 7623 | 0.409 | 0.1764 | No | ||
| 90 | BMP2K | 7685 | 0.399 | 0.1742 | No | ||
| 91 | NR5A1 | 7693 | 0.397 | 0.1750 | No | ||
| 92 | UTX | 7962 | 0.343 | 0.1615 | No | ||
| 93 | SIRT1 | 8112 | 0.315 | 0.1543 | No | ||
| 94 | DNAJB9 | 8648 | 0.214 | 0.1259 | No | ||
| 95 | TRPM7 | 8686 | 0.207 | 0.1246 | No | ||
| 96 | ALDH6A1 | 8915 | 0.170 | 0.1127 | No | ||
| 97 | NPM1 | 8937 | 0.166 | 0.1120 | No | ||
| 98 | NPTX1 | 8966 | 0.160 | 0.1110 | No | ||
| 99 | FOXF2 | 9038 | 0.147 | 0.1076 | No | ||
| 100 | ARMET | 9257 | 0.106 | 0.0961 | No | ||
| 101 | PHC3 | 9488 | 0.059 | 0.0838 | No | ||
| 102 | SLC38A5 | 9503 | 0.055 | 0.0832 | No | ||
| 103 | ODC1 | 9549 | 0.045 | 0.0809 | No | ||
| 104 | TIMM9 | 9635 | 0.024 | 0.0763 | No | ||
| 105 | FLT3 | 9819 | -0.016 | 0.0664 | No | ||
| 106 | ABCA1 | 9904 | -0.033 | 0.0620 | No | ||
| 107 | MRPL40 | 9979 | -0.055 | 0.0581 | No | ||
| 108 | BHLHB3 | 9995 | -0.058 | 0.0575 | No | ||
| 109 | HOXA11 | 9997 | -0.058 | 0.0576 | No | ||
| 110 | ADAMTS19 | 10227 | -0.107 | 0.0455 | No | ||
| 111 | NEUROD1 | 10311 | -0.126 | 0.0414 | No | ||
| 112 | HPS3 | 10507 | -0.169 | 0.0313 | No | ||
| 113 | TESK2 | 10584 | -0.183 | 0.0277 | No | ||
| 114 | LAMP1 | 10676 | -0.201 | 0.0234 | No | ||
| 115 | BMP4 | 11108 | -0.292 | 0.0009 | No | ||
| 116 | FGF14 | 11201 | -0.310 | -0.0032 | No | ||
| 117 | PAX6 | 11207 | -0.311 | -0.0026 | No | ||
| 118 | TGFB2 | 11211 | -0.313 | -0.0018 | No | ||
| 119 | CBARA1 | 11231 | -0.316 | -0.0019 | No | ||
| 120 | SNX5 | 11340 | -0.339 | -0.0068 | No | ||
| 121 | ARF6 | 11489 | -0.371 | -0.0137 | No | ||
| 122 | EGLN2 | 11601 | -0.395 | -0.0186 | No | ||
| 123 | NDUFA7 | 12306 | -0.558 | -0.0551 | No | ||
| 124 | GATA5 | 12320 | -0.562 | -0.0542 | No | ||
| 125 | STX6 | 12333 | -0.565 | -0.0532 | No | ||
| 126 | ETV4 | 12404 | -0.580 | -0.0552 | No | ||
| 127 | HOXB5 | 12480 | -0.599 | -0.0576 | No | ||
| 128 | ELAVL3 | 12621 | -0.632 | -0.0633 | No | ||
| 129 | SLCO1C1 | 12854 | -0.692 | -0.0738 | No | ||
| 130 | BDNF | 12902 | -0.703 | -0.0743 | No | ||
| 131 | PABPN1 | 13066 | -0.751 | -0.0809 | No | ||
| 132 | DCTN4 | 13091 | -0.757 | -0.0800 | No | ||
| 133 | JAG1 | 13173 | -0.779 | -0.0821 | No | ||
| 134 | RNF128 | 13442 | -0.851 | -0.0942 | No | ||
| 135 | B3GALT6 | 13443 | -0.851 | -0.0917 | No | ||
| 136 | MTUS1 | 13586 | -0.900 | -0.0967 | No | ||
| 137 | HOXA1 | 13771 | -0.955 | -0.1039 | No | ||
| 138 | RPA1 | 13806 | -0.965 | -0.1029 | No | ||
| 139 | ILF3 | 14335 | -1.146 | -0.1282 | No | ||
| 140 | SLC35A5 | 14355 | -1.152 | -0.1258 | No | ||
| 141 | TAC1 | 14458 | -1.189 | -0.1278 | No | ||
| 142 | DUSP7 | 14571 | -1.226 | -0.1303 | No | ||
| 143 | MNT | 14597 | -1.236 | -0.1280 | No | ||
| 144 | NRIP3 | 14637 | -1.250 | -0.1265 | No | ||
| 145 | DVL2 | 14755 | -1.299 | -0.1290 | No | ||
| 146 | SCYL1 | 14786 | -1.309 | -0.1268 | No | ||
| 147 | ESRRA | 14952 | -1.382 | -0.1317 | No | ||
| 148 | UBE2B | 15649 | -1.725 | -0.1643 | No | ||
| 149 | CBX5 | 15705 | -1.755 | -0.1622 | No | ||
| 150 | ARMCX3 | 15757 | -1.788 | -0.1597 | No | ||
| 151 | PDGFB | 15770 | -1.798 | -0.1550 | No | ||
| 152 | AKAP10 | 15845 | -1.846 | -0.1536 | No | ||
| 153 | HOXB7 | 15886 | -1.873 | -0.1503 | No | ||
| 154 | CSRP2BP | 16008 | -1.942 | -0.1511 | No | ||
| 155 | SMYD4 | 16061 | -1.973 | -0.1482 | No | ||
| 156 | IL15RA | 16258 | -2.145 | -0.1525 | No | ||
| 157 | LEF1 | 16397 | -2.266 | -0.1533 | No | ||
| 158 | ATF7IP | 16541 | -2.408 | -0.1540 | No | ||
| 159 | NIT1 | 16577 | -2.439 | -0.1487 | No | ||
| 160 | COMMD8 | 16599 | -2.457 | -0.1426 | No | ||
| 161 | UBXD1 | 16794 | -2.674 | -0.1453 | No | ||
| 162 | RTN4 | 16951 | -2.869 | -0.1453 | No | ||
| 163 | LZTS2 | 17184 | -3.207 | -0.1485 | No | ||
| 164 | VPS16 | 17345 | -3.485 | -0.1469 | No | ||
| 165 | AP1GBP1 | 17519 | -3.860 | -0.1450 | No | ||
| 166 | MAP2K6 | 17544 | -3.918 | -0.1348 | No | ||
| 167 | FXYD6 | 17853 | -4.680 | -0.1377 | No | ||
| 168 | EPC1 | 17925 | -4.900 | -0.1272 | No | ||
| 169 | BCL11B | 18037 | -5.272 | -0.1177 | No | ||
| 170 | KBTBD2 | 18041 | -5.284 | -0.1023 | No | ||
| 171 | BRD2 | 18095 | -5.480 | -0.0891 | No | ||
| 172 | RNF44 | 18193 | -5.987 | -0.0768 | No | ||
| 173 | ZFYVE26 | 18439 | -8.137 | -0.0661 | No | ||
| 174 | ISGF3G | 18468 | -8.565 | -0.0425 | No | ||
| 175 | HOXA7 | 18589 | -17.167 | 0.0015 | No |
