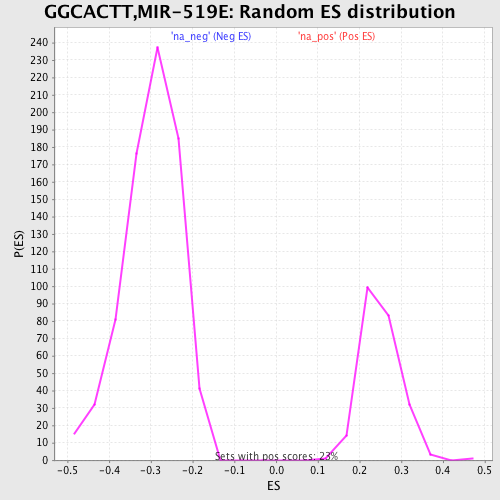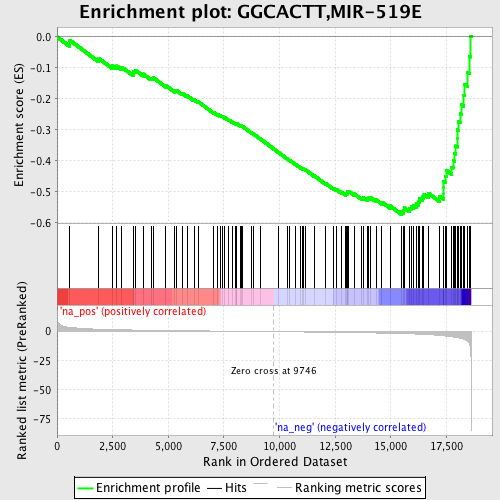
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GGCACTT,MIR-519E |
| Enrichment Score (ES) | -0.57301134 |
| Normalized Enrichment Score (NES) | -1.9241087 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.070574634 |
| FWER p-Value | 0.172 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DDX3X | 560 | 3.707 | -0.0099 | No | ||
| 2 | RPS6KA3 | 1863 | 2.086 | -0.0687 | No | ||
| 3 | CLOCK | 2497 | 1.756 | -0.0933 | No | ||
| 4 | C1GALT1 | 2676 | 1.679 | -0.0937 | No | ||
| 5 | NR4A2 | 2905 | 1.582 | -0.0973 | No | ||
| 6 | MMP19 | 3427 | 1.387 | -0.1178 | No | ||
| 7 | NPAS2 | 3437 | 1.384 | -0.1107 | No | ||
| 8 | VPS26A | 3503 | 1.364 | -0.1067 | No | ||
| 9 | DAZAP2 | 3867 | 1.248 | -0.1195 | No | ||
| 10 | MAP3K7 | 4229 | 1.146 | -0.1327 | No | ||
| 11 | ZNF148 | 4332 | 1.118 | -0.1320 | No | ||
| 12 | SEMA4G | 4886 | 0.986 | -0.1565 | No | ||
| 13 | WAC | 5287 | 0.887 | -0.1732 | No | ||
| 14 | SOX11 | 5371 | 0.868 | -0.1729 | No | ||
| 15 | CDK10 | 5617 | 0.817 | -0.1817 | No | ||
| 16 | DPYSL5 | 5846 | 0.769 | -0.1898 | No | ||
| 17 | PAFAH1B2 | 6155 | 0.707 | -0.2025 | No | ||
| 18 | MYST3 | 6354 | 0.664 | -0.2096 | No | ||
| 19 | ZBTB7A | 7029 | 0.524 | -0.2431 | No | ||
| 20 | EDG1 | 7205 | 0.490 | -0.2498 | No | ||
| 21 | QKI | 7336 | 0.465 | -0.2543 | No | ||
| 22 | LBP | 7412 | 0.449 | -0.2559 | No | ||
| 23 | ST8SIA2 | 7541 | 0.427 | -0.2604 | No | ||
| 24 | PTPRG | 7724 | 0.390 | -0.2681 | No | ||
| 25 | ARHGEF12 | 7879 | 0.359 | -0.2745 | No | ||
| 26 | DLL1 | 8022 | 0.332 | -0.2803 | No | ||
| 27 | MTMR4 | 8046 | 0.328 | -0.2798 | No | ||
| 28 | TBL1X | 8066 | 0.324 | -0.2790 | No | ||
| 29 | LPHN2 | 8231 | 0.295 | -0.2862 | No | ||
| 30 | BTBD7 | 8277 | 0.287 | -0.2871 | No | ||
| 31 | MMD | 8338 | 0.275 | -0.2888 | No | ||
| 32 | ZBTB4 | 8757 | 0.194 | -0.3103 | No | ||
| 33 | FGD1 | 8840 | 0.182 | -0.3138 | No | ||
| 34 | PTGFRN | 9162 | 0.123 | -0.3304 | No | ||
| 35 | PCYT1B | 9938 | -0.044 | -0.3720 | No | ||
| 36 | CORO7 | 10344 | -0.137 | -0.3931 | No | ||
| 37 | EIF2C1 | 10443 | -0.156 | -0.3976 | No | ||
| 38 | PTPN3 | 10707 | -0.206 | -0.4106 | No | ||
| 39 | TAGAP | 10947 | -0.257 | -0.4221 | No | ||
| 40 | EIF5A2 | 11044 | -0.278 | -0.4258 | No | ||
| 41 | MGEA5 | 11076 | -0.285 | -0.4259 | No | ||
| 42 | SCML2 | 11166 | -0.305 | -0.4290 | No | ||
| 43 | MYLK | 11553 | -0.385 | -0.4477 | No | ||
| 44 | RRM2 | 12069 | -0.498 | -0.4728 | No | ||
| 45 | NANOS1 | 12410 | -0.581 | -0.4880 | No | ||
| 46 | SP1 | 12543 | -0.615 | -0.4917 | No | ||
| 47 | CCNT2 | 12772 | -0.670 | -0.5004 | No | ||
| 48 | SON | 12965 | -0.720 | -0.5068 | No | ||
| 49 | FURIN | 13017 | -0.732 | -0.5055 | No | ||
| 50 | ARID4A | 13045 | -0.742 | -0.5029 | No | ||
| 51 | DKK1 | 13056 | -0.747 | -0.4993 | No | ||
| 52 | BCOR | 13104 | -0.761 | -0.4977 | No | ||
| 53 | SSFA2 | 13355 | -0.826 | -0.5067 | No | ||
| 54 | XRN1 | 13703 | -0.933 | -0.5203 | No | ||
| 55 | OSR1 | 13762 | -0.951 | -0.5182 | No | ||
| 56 | EFNB3 | 13968 | -1.019 | -0.5237 | No | ||
| 57 | NR2F2 | 14000 | -1.030 | -0.5197 | No | ||
| 58 | EPHA7 | 14068 | -1.053 | -0.5175 | No | ||
| 59 | BNIP2 | 14341 | -1.147 | -0.5259 | No | ||
| 60 | RBL2 | 14595 | -1.235 | -0.5328 | No | ||
| 61 | PTPRD | 14966 | -1.385 | -0.5452 | No | ||
| 62 | CXCR4 | 15483 | -1.636 | -0.5640 | Yes | ||
| 63 | PHF1 | 15575 | -1.683 | -0.5597 | Yes | ||
| 64 | IRF1 | 15595 | -1.695 | -0.5514 | Yes | ||
| 65 | PTEN | 15842 | -1.843 | -0.5546 | Yes | ||
| 66 | ZFHX4 | 15909 | -1.888 | -0.5478 | Yes | ||
| 67 | DDX3Y | 16020 | -1.949 | -0.5431 | Yes | ||
| 68 | FBXO21 | 16150 | -2.059 | -0.5387 | Yes | ||
| 69 | ZNF385 | 16254 | -2.139 | -0.5325 | Yes | ||
| 70 | MARK4 | 16276 | -2.162 | -0.5218 | Yes | ||
| 71 | MAT2B | 16409 | -2.276 | -0.5164 | Yes | ||
| 72 | AOF1 | 16489 | -2.352 | -0.5078 | Yes | ||
| 73 | SRPK1 | 16686 | -2.540 | -0.5044 | Yes | ||
| 74 | BICD2 | 17185 | -3.210 | -0.5137 | Yes | ||
| 75 | PPP2R2A | 17365 | -3.532 | -0.5040 | Yes | ||
| 76 | SERTAD2 | 17367 | -3.534 | -0.4847 | Yes | ||
| 77 | PERQ1 | 17370 | -3.536 | -0.4654 | Yes | ||
| 78 | RAB35 | 17463 | -3.721 | -0.4499 | Yes | ||
| 79 | ARID4B | 17499 | -3.822 | -0.4308 | Yes | ||
| 80 | UBE3C | 17742 | -4.355 | -0.4200 | Yes | ||
| 81 | CCNG2 | 17799 | -4.518 | -0.3982 | Yes | ||
| 82 | PAFAH1B1 | 17867 | -4.737 | -0.3758 | Yes | ||
| 83 | MKRN1 | 17922 | -4.891 | -0.3519 | Yes | ||
| 84 | PLAGL2 | 17986 | -5.060 | -0.3275 | Yes | ||
| 85 | SPATA2 | 17990 | -5.095 | -0.2998 | Yes | ||
| 86 | BCL11B | 18037 | -5.272 | -0.2733 | Yes | ||
| 87 | ZIC4 | 18117 | -5.585 | -0.2469 | Yes | ||
| 88 | RAB5B | 18167 | -5.832 | -0.2176 | Yes | ||
| 89 | MARK2 | 18285 | -6.557 | -0.1879 | Yes | ||
| 90 | NEDD4L | 18304 | -6.734 | -0.1519 | Yes | ||
| 91 | ARHGEF5 | 18448 | -8.219 | -0.1145 | Yes | ||
| 92 | ARL4C | 18528 | -10.209 | -0.0628 | Yes | ||
| 93 | EHD3 | 18567 | -12.294 | 0.0026 | Yes |