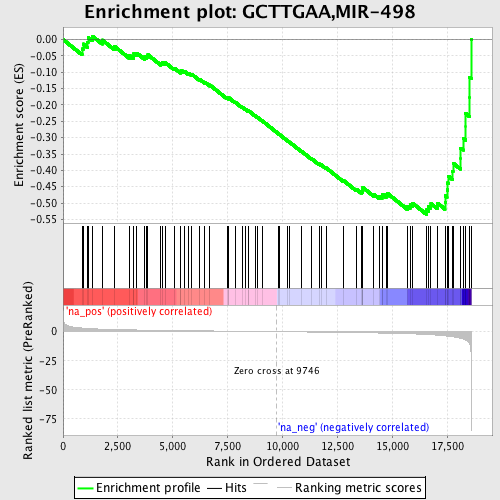
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GCTTGAA,MIR-498 |
| Enrichment Score (ES) | -0.5349947 |
| Normalized Enrichment Score (NES) | -1.7732184 |
| Nominal p-value | 0.0026041667 |
| FDR q-value | 0.103861175 |
| FWER p-Value | 0.602 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RNF125 | 865 | 3.132 | -0.0285 | No | ||
| 2 | RNF11 | 912 | 3.053 | -0.0134 | No | ||
| 3 | TFRC | 1111 | 2.779 | -0.0080 | No | ||
| 4 | PACSIN2 | 1138 | 2.749 | 0.0065 | No | ||
| 5 | FUSIP1 | 1332 | 2.512 | 0.0107 | No | ||
| 6 | NARG1 | 1782 | 2.135 | -0.0012 | No | ||
| 7 | CCNE1 | 2346 | 1.829 | -0.0210 | No | ||
| 8 | DTNA | 3017 | 1.537 | -0.0483 | No | ||
| 9 | OLIG3 | 3217 | 1.469 | -0.0505 | No | ||
| 10 | PHF6 | 3224 | 1.467 | -0.0423 | No | ||
| 11 | RUNX2 | 3357 | 1.416 | -0.0413 | No | ||
| 12 | PDIA6 | 3712 | 1.295 | -0.0529 | No | ||
| 13 | KPNB1 | 3811 | 1.264 | -0.0508 | No | ||
| 14 | PLEKHC1 | 3866 | 1.248 | -0.0465 | No | ||
| 15 | NAP1L3 | 4452 | 1.088 | -0.0718 | No | ||
| 16 | WT1 | 4512 | 1.075 | -0.0688 | No | ||
| 17 | COL1A1 | 4663 | 1.040 | -0.0708 | No | ||
| 18 | PALM2 | 5069 | 0.943 | -0.0872 | No | ||
| 19 | ARRDC3 | 5353 | 0.872 | -0.0975 | No | ||
| 20 | SOX11 | 5371 | 0.868 | -0.0934 | No | ||
| 21 | GJA1 | 5530 | 0.833 | -0.0971 | No | ||
| 22 | CACNA2D3 | 5731 | 0.793 | -0.1033 | No | ||
| 23 | MORF4L2 | 5856 | 0.768 | -0.1055 | No | ||
| 24 | PIK3R1 | 6233 | 0.693 | -0.1218 | No | ||
| 25 | CAMK2G | 6454 | 0.643 | -0.1299 | No | ||
| 26 | KCND2 | 6680 | 0.598 | -0.1386 | No | ||
| 27 | MAB21L1 | 7474 | 0.439 | -0.1789 | No | ||
| 28 | CPNE8 | 7535 | 0.427 | -0.1796 | No | ||
| 29 | ST8SIA2 | 7541 | 0.427 | -0.1774 | No | ||
| 30 | EML4 | 7840 | 0.368 | -0.1914 | No | ||
| 31 | DHX35 | 8158 | 0.306 | -0.2067 | No | ||
| 32 | DUSP4 | 8297 | 0.283 | -0.2125 | No | ||
| 33 | RBPMS | 8434 | 0.257 | -0.2184 | No | ||
| 34 | ITSN1 | 8453 | 0.255 | -0.2179 | No | ||
| 35 | FANCA | 8752 | 0.195 | -0.2328 | No | ||
| 36 | UBE2J1 | 8880 | 0.176 | -0.2386 | No | ||
| 37 | EXOC5 | 9107 | 0.133 | -0.2501 | No | ||
| 38 | FLRT2 | 9823 | -0.017 | -0.2885 | No | ||
| 39 | WDR26 | 9882 | -0.029 | -0.2915 | No | ||
| 40 | MAPRE1 | 10217 | -0.105 | -0.3089 | No | ||
| 41 | SPRED1 | 10327 | -0.132 | -0.3140 | No | ||
| 42 | LRP1B | 10879 | -0.245 | -0.3423 | No | ||
| 43 | CBX4 | 11314 | -0.334 | -0.3638 | No | ||
| 44 | SIX4 | 11664 | -0.410 | -0.3803 | No | ||
| 45 | CRH | 11754 | -0.429 | -0.3826 | No | ||
| 46 | PAM | 11990 | -0.483 | -0.3925 | No | ||
| 47 | CCNT2 | 12772 | -0.670 | -0.4307 | No | ||
| 48 | SSFA2 | 13355 | -0.826 | -0.4574 | No | ||
| 49 | SOCS3 | 13595 | -0.902 | -0.4650 | No | ||
| 50 | CHODL | 13632 | -0.912 | -0.4617 | No | ||
| 51 | PPP3CA | 13649 | -0.916 | -0.4573 | No | ||
| 52 | DICER1 | 13657 | -0.918 | -0.4523 | No | ||
| 53 | PKNOX2 | 14155 | -1.082 | -0.4729 | No | ||
| 54 | MEIS1 | 14412 | -1.172 | -0.4799 | No | ||
| 55 | ATPAF1 | 14537 | -1.215 | -0.4796 | No | ||
| 56 | CAPZA2 | 14565 | -1.226 | -0.4739 | No | ||
| 57 | RNF149 | 14722 | -1.286 | -0.4749 | No | ||
| 58 | DCBLD2 | 14793 | -1.312 | -0.4711 | No | ||
| 59 | YPEL2 | 15699 | -1.750 | -0.5098 | No | ||
| 60 | RAB6A | 15814 | -1.824 | -0.5054 | No | ||
| 61 | PDE4B | 15923 | -1.895 | -0.5003 | No | ||
| 62 | RAB11A | 16568 | -2.430 | -0.5209 | Yes | ||
| 63 | GRIA2 | 16654 | -2.515 | -0.5110 | Yes | ||
| 64 | KLF12 | 16764 | -2.638 | -0.5016 | Yes | ||
| 65 | HIPK1 | 17061 | -3.001 | -0.5002 | Yes | ||
| 66 | NCOA1 | 17406 | -3.604 | -0.4979 | Yes | ||
| 67 | ABTB1 | 17421 | -3.622 | -0.4777 | Yes | ||
| 68 | MECP2 | 17498 | -3.820 | -0.4597 | Yes | ||
| 69 | IRF2 | 17520 | -3.862 | -0.4385 | Yes | ||
| 70 | CTNS | 17554 | -3.930 | -0.4176 | Yes | ||
| 71 | CUGBP1 | 17752 | -4.386 | -0.4028 | Yes | ||
| 72 | RAB22A | 17775 | -4.485 | -0.3780 | Yes | ||
| 73 | ZFP36L1 | 18112 | -5.581 | -0.3639 | Yes | ||
| 74 | ZIC4 | 18117 | -5.585 | -0.3318 | Yes | ||
| 75 | SS18L1 | 18232 | -6.212 | -0.3020 | Yes | ||
| 76 | HBP1 | 18344 | -7.095 | -0.2669 | Yes | ||
| 77 | SNRK | 18346 | -7.103 | -0.2259 | Yes | ||
| 78 | ARL4C | 18528 | -10.209 | -0.1766 | Yes | ||
| 79 | PURG | 18533 | -10.451 | -0.1164 | Yes | ||
| 80 | BACH2 | 18601 | -20.879 | 0.0008 | Yes |
