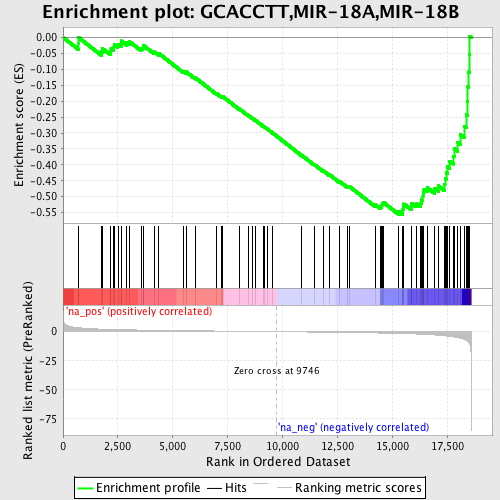
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GCACCTT,MIR-18A,MIR-18B |
| Enrichment Score (ES) | -0.55581796 |
| Normalized Enrichment Score (NES) | -1.8344086 |
| Nominal p-value | 0.002754821 |
| FDR q-value | 0.08393516 |
| FWER p-Value | 0.408 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IGF1 | 712 | 3.391 | -0.0195 | No | ||
| 2 | TRIM2 | 717 | 3.383 | -0.0009 | No | ||
| 3 | NR3C1 | 1754 | 2.156 | -0.0448 | No | ||
| 4 | SIM2 | 1787 | 2.131 | -0.0346 | No | ||
| 5 | MESP1 | 2164 | 1.920 | -0.0442 | No | ||
| 6 | ARL15 | 2181 | 1.912 | -0.0344 | No | ||
| 7 | AKR1D1 | 2306 | 1.844 | -0.0309 | No | ||
| 8 | PHF19 | 2319 | 1.840 | -0.0213 | No | ||
| 9 | CDC42 | 2501 | 1.755 | -0.0213 | No | ||
| 10 | NAV1 | 2647 | 1.692 | -0.0197 | No | ||
| 11 | ALCAM | 2660 | 1.685 | -0.0109 | No | ||
| 12 | ESR1 | 2907 | 1.581 | -0.0154 | No | ||
| 13 | TEX2 | 3040 | 1.529 | -0.0140 | No | ||
| 14 | SOCS5 | 3558 | 1.346 | -0.0344 | No | ||
| 15 | PIAS3 | 3642 | 1.315 | -0.0316 | No | ||
| 16 | MAN1A2 | 3645 | 1.314 | -0.0243 | No | ||
| 17 | PRICKLE2 | 4158 | 1.164 | -0.0455 | No | ||
| 18 | BHLHB5 | 4364 | 1.111 | -0.0504 | No | ||
| 19 | ASXL2 | 5496 | 0.840 | -0.1067 | No | ||
| 20 | CTGF | 5609 | 0.819 | -0.1082 | No | ||
| 21 | DPP10 | 6025 | 0.731 | -0.1265 | No | ||
| 22 | BTG3 | 6999 | 0.530 | -0.1760 | No | ||
| 23 | RHOT1 | 7228 | 0.486 | -0.1856 | No | ||
| 24 | RAB11FIP2 | 7247 | 0.483 | -0.1839 | No | ||
| 25 | LIN28 | 8035 | 0.329 | -0.2245 | No | ||
| 26 | HSF2 | 8449 | 0.255 | -0.2453 | No | ||
| 27 | RABGAP1 | 8643 | 0.214 | -0.2546 | No | ||
| 28 | ZBTB4 | 8757 | 0.194 | -0.2596 | No | ||
| 29 | TARDBP | 9136 | 0.129 | -0.2792 | No | ||
| 30 | PTGFRN | 9162 | 0.123 | -0.2799 | No | ||
| 31 | CTDSPL | 9324 | 0.092 | -0.2881 | No | ||
| 32 | PURB | 9550 | 0.045 | -0.3000 | No | ||
| 33 | PRKACB | 10855 | -0.239 | -0.3690 | No | ||
| 34 | KIT | 11438 | -0.360 | -0.3984 | No | ||
| 35 | WSB1 | 11858 | -0.453 | -0.4184 | No | ||
| 36 | SH3BP4 | 12145 | -0.516 | -0.4310 | No | ||
| 37 | SAR1A | 12618 | -0.631 | -0.4529 | No | ||
| 38 | SON | 12965 | -0.720 | -0.4676 | No | ||
| 39 | FRMD4A | 13043 | -0.741 | -0.4676 | No | ||
| 40 | MEF2D | 14230 | -1.112 | -0.5254 | No | ||
| 41 | GLRB | 14464 | -1.193 | -0.5313 | No | ||
| 42 | HIF1A | 14529 | -1.213 | -0.5280 | No | ||
| 43 | MDGA1 | 14536 | -1.215 | -0.5216 | No | ||
| 44 | GCLC | 14615 | -1.243 | -0.5189 | No | ||
| 45 | CLASP2 | 15299 | -1.538 | -0.5471 | Yes | ||
| 46 | FCHSD2 | 15461 | -1.625 | -0.5468 | Yes | ||
| 47 | EHMT1 | 15488 | -1.638 | -0.5391 | Yes | ||
| 48 | ETV6 | 15503 | -1.645 | -0.5307 | Yes | ||
| 49 | CRIM1 | 15531 | -1.657 | -0.5229 | Yes | ||
| 50 | STK4 | 15864 | -1.857 | -0.5305 | Yes | ||
| 51 | NR1H2 | 15893 | -1.877 | -0.5215 | Yes | ||
| 52 | SMAD2 | 16091 | -1.995 | -0.5210 | Yes | ||
| 53 | XYLT2 | 16297 | -2.182 | -0.5199 | Yes | ||
| 54 | TRIB2 | 16336 | -2.218 | -0.5096 | Yes | ||
| 55 | RAB5C | 16399 | -2.270 | -0.5003 | Yes | ||
| 56 | TRIOBP | 16404 | -2.274 | -0.4879 | Yes | ||
| 57 | RUNX1 | 16446 | -2.312 | -0.4772 | Yes | ||
| 58 | JARID1B | 16590 | -2.450 | -0.4713 | Yes | ||
| 59 | FNBP1 | 16941 | -2.853 | -0.4743 | Yes | ||
| 60 | ADD3 | 17108 | -3.081 | -0.4661 | Yes | ||
| 61 | PERQ1 | 17370 | -3.536 | -0.4605 | Yes | ||
| 62 | NCOA1 | 17406 | -3.604 | -0.4423 | Yes | ||
| 63 | CLK2 | 17470 | -3.735 | -0.4249 | Yes | ||
| 64 | IRF2 | 17520 | -3.862 | -0.4061 | Yes | ||
| 65 | ATXN1 | 17611 | -4.029 | -0.3885 | Yes | ||
| 66 | NFAT5 | 17779 | -4.490 | -0.3725 | Yes | ||
| 67 | AEBP2 | 17820 | -4.582 | -0.3492 | Yes | ||
| 68 | ANKRD50 | 17989 | -5.072 | -0.3300 | Yes | ||
| 69 | ZFP36L1 | 18112 | -5.581 | -0.3055 | Yes | ||
| 70 | PARP6 | 18278 | -6.517 | -0.2781 | Yes | ||
| 71 | NEDD9 | 18379 | -7.388 | -0.2424 | Yes | ||
| 72 | PACSIN1 | 18434 | -8.081 | -0.2003 | Yes | ||
| 73 | ARHGEF5 | 18448 | -8.219 | -0.1553 | Yes | ||
| 74 | SNURF | 18471 | -8.632 | -0.1084 | Yes | ||
| 75 | GAB1 | 18529 | -10.215 | -0.0546 | Yes | ||
| 76 | PHF2 | 18539 | -10.641 | 0.0042 | Yes |
