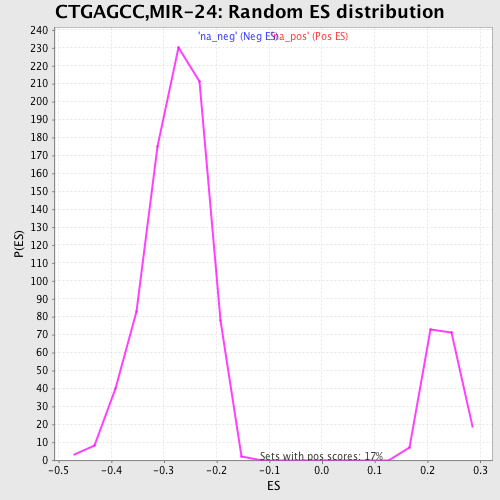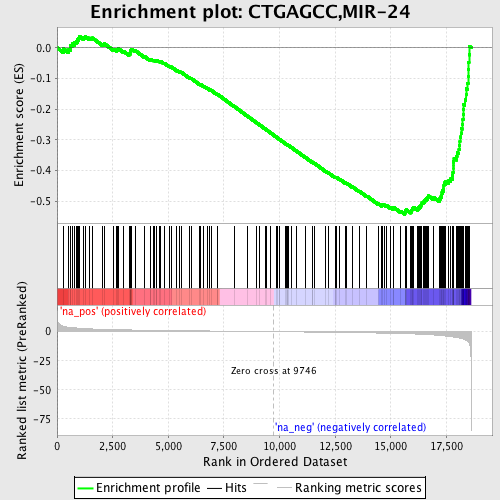
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CTGAGCC,MIR-24 |
| Enrichment Score (ES) | -0.5431206 |
| Normalized Enrichment Score (NES) | -1.9452257 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.08617126 |
| FWER p-Value | 0.143 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ABCB9 | 281 | 4.656 | -0.0029 | No | ||
| 2 | RNF138 | 526 | 3.786 | -0.0061 | No | ||
| 3 | DOCK3 | 598 | 3.601 | -0.0004 | No | ||
| 4 | PPM1G | 604 | 3.591 | 0.0088 | No | ||
| 5 | MAFG | 674 | 3.447 | 0.0142 | No | ||
| 6 | AARS | 786 | 3.263 | 0.0168 | No | ||
| 7 | GPX3 | 858 | 3.144 | 0.0212 | No | ||
| 8 | RNF11 | 912 | 3.053 | 0.0264 | No | ||
| 9 | SEMA6B | 969 | 2.959 | 0.0312 | No | ||
| 10 | CCT3 | 984 | 2.944 | 0.0383 | No | ||
| 11 | NEK4 | 1189 | 2.677 | 0.0343 | No | ||
| 12 | KPNA3 | 1257 | 2.592 | 0.0375 | No | ||
| 13 | PDGFRA | 1473 | 2.373 | 0.0321 | No | ||
| 14 | PER1 | 1581 | 2.278 | 0.0324 | No | ||
| 15 | ING5 | 2052 | 1.985 | 0.0121 | No | ||
| 16 | B3GNT6 | 2121 | 1.940 | 0.0136 | No | ||
| 17 | HAP1 | 2544 | 1.734 | -0.0047 | No | ||
| 18 | MATR3 | 2684 | 1.674 | -0.0078 | No | ||
| 19 | XPO4 | 2722 | 1.658 | -0.0054 | No | ||
| 20 | TOP1 | 2747 | 1.646 | -0.0024 | No | ||
| 21 | FGFR3 | 2991 | 1.548 | -0.0114 | No | ||
| 22 | JUB | 3233 | 1.464 | -0.0206 | No | ||
| 23 | RNF150 | 3280 | 1.444 | -0.0193 | No | ||
| 24 | NDST1 | 3293 | 1.442 | -0.0161 | No | ||
| 25 | KLHL1 | 3296 | 1.441 | -0.0124 | No | ||
| 26 | PCDH10 | 3310 | 1.434 | -0.0093 | No | ||
| 27 | PRSS8 | 3328 | 1.427 | -0.0065 | No | ||
| 28 | TAF5 | 3349 | 1.420 | -0.0038 | No | ||
| 29 | INSIG1 | 3515 | 1.361 | -0.0092 | No | ||
| 30 | TRIM33 | 3925 | 1.228 | -0.0281 | No | ||
| 31 | RAP1B | 4190 | 1.155 | -0.0393 | No | ||
| 32 | FBXL19 | 4199 | 1.153 | -0.0367 | No | ||
| 33 | FST | 4314 | 1.125 | -0.0399 | No | ||
| 34 | BHLHB5 | 4364 | 1.111 | -0.0396 | No | ||
| 35 | LENG4 | 4469 | 1.084 | -0.0424 | No | ||
| 36 | MAPK7 | 4474 | 1.083 | -0.0397 | No | ||
| 37 | NEK9 | 4594 | 1.057 | -0.0434 | No | ||
| 38 | TNFRSF19 | 4664 | 1.040 | -0.0444 | No | ||
| 39 | CDV3 | 4810 | 1.003 | -0.0496 | No | ||
| 40 | BCL2L11 | 5035 | 0.951 | -0.0592 | No | ||
| 41 | TCF2 | 5136 | 0.926 | -0.0622 | No | ||
| 42 | EDA | 5384 | 0.864 | -0.0733 | No | ||
| 43 | PIGS | 5504 | 0.838 | -0.0775 | No | ||
| 44 | CMTM4 | 5582 | 0.825 | -0.0795 | No | ||
| 45 | NELL2 | 5936 | 0.752 | -0.0966 | No | ||
| 46 | RAP2C | 6022 | 0.732 | -0.0993 | No | ||
| 47 | RASGRP4 | 6402 | 0.653 | -0.1181 | No | ||
| 48 | PTGER4 | 6462 | 0.641 | -0.1196 | No | ||
| 49 | SLC19A2 | 6589 | 0.617 | -0.1248 | No | ||
| 50 | DLC1 | 6753 | 0.581 | -0.1321 | No | ||
| 51 | STC2 | 6866 | 0.559 | -0.1367 | No | ||
| 52 | SMCR8 | 6930 | 0.544 | -0.1387 | No | ||
| 53 | EDG1 | 7205 | 0.490 | -0.1522 | No | ||
| 54 | RAP1A | 7210 | 0.489 | -0.1511 | No | ||
| 55 | BSN | 7975 | 0.340 | -0.1916 | No | ||
| 56 | TRPM6 | 8551 | 0.233 | -0.2221 | No | ||
| 57 | CALCR | 8964 | 0.160 | -0.2440 | No | ||
| 58 | EXOC5 | 9107 | 0.133 | -0.2514 | No | ||
| 59 | MMP16 | 9346 | 0.087 | -0.2640 | No | ||
| 60 | AMMECR1 | 9404 | 0.078 | -0.2669 | No | ||
| 61 | AMOTL2 | 9595 | 0.036 | -0.2771 | No | ||
| 62 | GPC4 | 9840 | -0.020 | -0.2903 | No | ||
| 63 | PURA | 9884 | -0.030 | -0.2925 | No | ||
| 64 | RARG | 9975 | -0.054 | -0.2973 | No | ||
| 65 | LYPLA2 | 9996 | -0.058 | -0.2982 | No | ||
| 66 | ARID5B | 10261 | -0.115 | -0.3122 | No | ||
| 67 | NEUROD1 | 10311 | -0.126 | -0.3145 | No | ||
| 68 | IGFBP5 | 10362 | -0.140 | -0.3169 | No | ||
| 69 | YBX2 | 10371 | -0.141 | -0.3169 | No | ||
| 70 | EPHA8 | 10402 | -0.148 | -0.3182 | No | ||
| 71 | PTPN9 | 10531 | -0.175 | -0.3246 | No | ||
| 72 | MAP3K7IP2 | 10766 | -0.219 | -0.3367 | No | ||
| 73 | SCML2 | 11166 | -0.305 | -0.3575 | No | ||
| 74 | APBA1 | 11458 | -0.365 | -0.3723 | No | ||
| 75 | TMEM9 | 11470 | -0.366 | -0.3720 | No | ||
| 76 | LRFN2 | 11570 | -0.388 | -0.3763 | No | ||
| 77 | CDX2 | 12059 | -0.496 | -0.4014 | No | ||
| 78 | CLCN6 | 12202 | -0.531 | -0.4077 | No | ||
| 79 | APOA5 | 12505 | -0.604 | -0.4225 | No | ||
| 80 | SFXN5 | 12522 | -0.609 | -0.4217 | No | ||
| 81 | SP1 | 12543 | -0.615 | -0.4212 | No | ||
| 82 | ADCY9 | 12714 | -0.658 | -0.4287 | No | ||
| 83 | PHC1 | 12954 | -0.717 | -0.4397 | No | ||
| 84 | FURIN | 13017 | -0.732 | -0.4411 | No | ||
| 85 | COL11A2 | 13275 | -0.803 | -0.4529 | No | ||
| 86 | ACVR1B | 13573 | -0.893 | -0.4667 | No | ||
| 87 | DLGAP4 | 13924 | -1.001 | -0.4830 | No | ||
| 88 | MMP14 | 14425 | -1.177 | -0.5069 | No | ||
| 89 | MNT | 14597 | -1.236 | -0.5129 | No | ||
| 90 | PER2 | 14636 | -1.250 | -0.5117 | No | ||
| 91 | RASA1 | 14704 | -1.279 | -0.5119 | No | ||
| 92 | DCBLD2 | 14793 | -1.312 | -0.5132 | No | ||
| 93 | PTPRD | 14966 | -1.385 | -0.5189 | No | ||
| 94 | PAK4 | 15098 | -1.444 | -0.5222 | No | ||
| 95 | WDTC1 | 15109 | -1.448 | -0.5189 | No | ||
| 96 | WDR23 | 15429 | -1.609 | -0.5319 | No | ||
| 97 | CACNB1 | 15637 | -1.717 | -0.5386 | Yes | ||
| 98 | DNAJB12 | 15641 | -1.721 | -0.5342 | Yes | ||
| 99 | SUV420H2 | 15656 | -1.729 | -0.5304 | Yes | ||
| 100 | SBK1 | 15689 | -1.743 | -0.5275 | Yes | ||
| 101 | GRIA3 | 15891 | -1.875 | -0.5334 | Yes | ||
| 102 | PLK3 | 15928 | -1.897 | -0.5304 | Yes | ||
| 103 | ADAM19 | 15952 | -1.908 | -0.5266 | Yes | ||
| 104 | SP6 | 15979 | -1.922 | -0.5229 | Yes | ||
| 105 | HIC2 | 16009 | -1.942 | -0.5193 | Yes | ||
| 106 | INPP5B | 16215 | -2.106 | -0.5249 | Yes | ||
| 107 | ATP6V0A2 | 16231 | -2.120 | -0.5201 | Yes | ||
| 108 | MARK4 | 16276 | -2.162 | -0.5167 | Yes | ||
| 109 | DHX30 | 16349 | -2.226 | -0.5148 | Yes | ||
| 110 | SYP | 16363 | -2.240 | -0.5095 | Yes | ||
| 111 | RAB5C | 16399 | -2.270 | -0.5054 | Yes | ||
| 112 | RNF2 | 16454 | -2.318 | -0.5022 | Yes | ||
| 113 | KCNK2 | 16503 | -2.366 | -0.4986 | Yes | ||
| 114 | CSK | 16538 | -2.400 | -0.4941 | Yes | ||
| 115 | PRKRIP1 | 16601 | -2.460 | -0.4909 | Yes | ||
| 116 | H2AFX | 16660 | -2.521 | -0.4874 | Yes | ||
| 117 | PTPRF | 16694 | -2.553 | -0.4824 | Yes | ||
| 118 | BCL2L2 | 16936 | -2.850 | -0.4880 | Yes | ||
| 119 | POGZ | 17173 | -3.185 | -0.4923 | Yes | ||
| 120 | HIP1R | 17217 | -3.254 | -0.4861 | Yes | ||
| 121 | MOCS1 | 17271 | -3.339 | -0.4801 | Yes | ||
| 122 | CNTFR | 17297 | -3.389 | -0.4725 | Yes | ||
| 123 | PLOD2 | 17321 | -3.428 | -0.4647 | Yes | ||
| 124 | OGT | 17379 | -3.550 | -0.4584 | Yes | ||
| 125 | ACBD4 | 17384 | -3.557 | -0.4492 | Yes | ||
| 126 | SESN1 | 17393 | -3.569 | -0.4402 | Yes | ||
| 127 | CLK2 | 17470 | -3.735 | -0.4344 | Yes | ||
| 128 | MAGI1 | 17608 | -4.021 | -0.4312 | Yes | ||
| 129 | GBA2 | 17698 | -4.255 | -0.4248 | Yes | ||
| 130 | PIM1 | 17761 | -4.415 | -0.4165 | Yes | ||
| 131 | PIM2 | 17790 | -4.508 | -0.4061 | Yes | ||
| 132 | MXI1 | 17802 | -4.527 | -0.3947 | Yes | ||
| 133 | PLCL2 | 17806 | -4.544 | -0.3828 | Yes | ||
| 134 | RANBP10 | 17810 | -4.553 | -0.3710 | Yes | ||
| 135 | ALS2CR2 | 17838 | -4.629 | -0.3602 | Yes | ||
| 136 | VAV1 | 17943 | -4.944 | -0.3528 | Yes | ||
| 137 | ZXDA | 17977 | -5.033 | -0.3412 | Yes | ||
| 138 | RHBDL1 | 18062 | -5.355 | -0.3316 | Yes | ||
| 139 | PSAP | 18075 | -5.395 | -0.3180 | Yes | ||
| 140 | RGL2 | 18106 | -5.554 | -0.3050 | Yes | ||
| 141 | MIDN | 18111 | -5.577 | -0.2904 | Yes | ||
| 142 | RASSF2 | 18154 | -5.765 | -0.2775 | Yes | ||
| 143 | RAB5B | 18167 | -5.832 | -0.2627 | Yes | ||
| 144 | MEN1 | 18204 | -6.041 | -0.2487 | Yes | ||
| 145 | TOB2 | 18239 | -6.235 | -0.2340 | Yes | ||
| 146 | KIF21B | 18252 | -6.347 | -0.2179 | Yes | ||
| 147 | SEMA4A | 18266 | -6.393 | -0.2017 | Yes | ||
| 148 | PARP6 | 18278 | -6.517 | -0.1851 | Yes | ||
| 149 | SSH2 | 18334 | -6.983 | -0.1696 | Yes | ||
| 150 | NEDD9 | 18379 | -7.388 | -0.1524 | Yes | ||
| 151 | PIP5K1B | 18388 | -7.474 | -0.1331 | Yes | ||
| 152 | ARHGEF5 | 18448 | -8.219 | -0.1146 | Yes | ||
| 153 | CDKN1B | 18474 | -8.692 | -0.0929 | Yes | ||
| 154 | TRPC4AP | 18483 | -8.827 | -0.0700 | Yes | ||
| 155 | SLC12A6 | 18499 | -9.098 | -0.0468 | Yes | ||
| 156 | DUSP16 | 18515 | -9.556 | -0.0223 | Yes | ||
| 157 | RAB6IP1 | 18535 | -10.499 | 0.0044 | Yes |