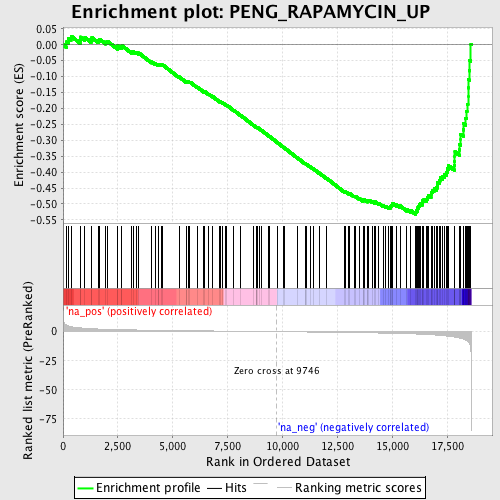
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | PENG_RAPAMYCIN_UP |
| Enrichment Score (ES) | -0.532936 |
| Normalized Enrichment Score (NES) | -1.874123 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.118895315 |
| FWER p-Value | 0.896 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EIF3S6 | 160 | 5.549 | 0.0089 | No | ||
| 2 | DRAP1 | 250 | 4.858 | 0.0195 | No | ||
| 3 | MTMR2 | 361 | 4.289 | 0.0272 | No | ||
| 4 | QSCN6 | 777 | 3.275 | 0.0151 | No | ||
| 5 | AP4M1 | 796 | 3.247 | 0.0244 | No | ||
| 6 | CAMK1 | 992 | 2.935 | 0.0232 | No | ||
| 7 | PLTP | 1276 | 2.562 | 0.0160 | No | ||
| 8 | MYC | 1312 | 2.533 | 0.0221 | No | ||
| 9 | LY86 | 1618 | 2.256 | 0.0128 | No | ||
| 10 | RGS13 | 1666 | 2.222 | 0.0173 | No | ||
| 11 | IDH2 | 1945 | 2.041 | 0.0087 | No | ||
| 12 | ACADVL | 2013 | 2.004 | 0.0114 | No | ||
| 13 | ALDH2 | 2482 | 1.761 | -0.0083 | No | ||
| 14 | CLOCK | 2497 | 1.756 | -0.0035 | No | ||
| 15 | DDX46 | 2669 | 1.683 | -0.0074 | No | ||
| 16 | CD22 | 2682 | 1.675 | -0.0027 | No | ||
| 17 | TOP2A | 3129 | 1.499 | -0.0221 | No | ||
| 18 | CD48 | 3202 | 1.473 | -0.0214 | No | ||
| 19 | CCNG1 | 3334 | 1.425 | -0.0239 | No | ||
| 20 | TK1 | 3446 | 1.381 | -0.0255 | No | ||
| 21 | HAGH | 4034 | 1.195 | -0.0535 | No | ||
| 22 | GNB3 | 4213 | 1.150 | -0.0595 | No | ||
| 23 | TCF20 | 4339 | 1.116 | -0.0627 | No | ||
| 24 | CFLAR | 4367 | 1.109 | -0.0607 | No | ||
| 25 | HMMR | 4476 | 1.083 | -0.0631 | No | ||
| 26 | FKBP8 | 4542 | 1.066 | -0.0632 | No | ||
| 27 | HIST1H3H | 5291 | 0.886 | -0.1009 | No | ||
| 28 | RQCD1 | 5613 | 0.818 | -0.1157 | No | ||
| 29 | SPOCK2 | 5630 | 0.814 | -0.1140 | No | ||
| 30 | IFIT1 | 5704 | 0.798 | -0.1154 | No | ||
| 31 | ASCL1 | 5781 | 0.783 | -0.1170 | No | ||
| 32 | NOTCH4 | 6125 | 0.713 | -0.1333 | No | ||
| 33 | RASGRP3 | 6416 | 0.651 | -0.1469 | No | ||
| 34 | HIST1H2BK | 6424 | 0.650 | -0.1453 | No | ||
| 35 | SURF1 | 6642 | 0.606 | -0.1551 | No | ||
| 36 | TP53 | 6813 | 0.567 | -0.1625 | No | ||
| 37 | RTN2 | 7138 | 0.504 | -0.1784 | No | ||
| 38 | HIST1H2BN | 7178 | 0.495 | -0.1789 | No | ||
| 39 | BNIP3L | 7263 | 0.480 | -0.1820 | No | ||
| 40 | HMG20B | 7400 | 0.450 | -0.1879 | No | ||
| 41 | MDM4 | 7454 | 0.442 | -0.1894 | No | ||
| 42 | FUCA1 | 7780 | 0.380 | -0.2058 | No | ||
| 43 | CIRBP | 8079 | 0.321 | -0.2209 | No | ||
| 44 | GPX6 | 8671 | 0.209 | -0.2522 | No | ||
| 45 | CDT1 | 8800 | 0.188 | -0.2585 | No | ||
| 46 | RBMS3 | 8829 | 0.184 | -0.2594 | No | ||
| 47 | CENPF | 8830 | 0.184 | -0.2589 | No | ||
| 48 | UBE2J1 | 8880 | 0.176 | -0.2610 | No | ||
| 49 | MS4A2 | 8928 | 0.167 | -0.2630 | No | ||
| 50 | SASH1 | 9060 | 0.143 | -0.2696 | No | ||
| 51 | SSTR2 | 9377 | 0.082 | -0.2865 | No | ||
| 52 | PTPRH | 9387 | 0.080 | -0.2867 | No | ||
| 53 | FAIM2 | 9768 | -0.006 | -0.3072 | No | ||
| 54 | MUSK | 10041 | -0.067 | -0.3217 | No | ||
| 55 | CRHR2 | 10088 | -0.076 | -0.3240 | No | ||
| 56 | ENPP2 | 10689 | -0.204 | -0.3558 | No | ||
| 57 | POU2AF1 | 11033 | -0.276 | -0.3735 | No | ||
| 58 | ARHGAP1 | 11045 | -0.278 | -0.3732 | No | ||
| 59 | GRINL1A | 11090 | -0.288 | -0.3747 | No | ||
| 60 | DAAM1 | 11278 | -0.326 | -0.3838 | No | ||
| 61 | MS4A1 | 11396 | -0.350 | -0.3890 | No | ||
| 62 | DMBT1 | 11671 | -0.411 | -0.4025 | No | ||
| 63 | MEG3 | 12019 | -0.488 | -0.4197 | No | ||
| 64 | CBLB | 12833 | -0.686 | -0.4616 | No | ||
| 65 | ERCC1 | 12864 | -0.694 | -0.4610 | No | ||
| 66 | JAK1 | 13010 | -0.731 | -0.4665 | No | ||
| 67 | BCKDHA | 13063 | -0.750 | -0.4669 | No | ||
| 68 | PDLIM1 | 13266 | -0.801 | -0.4753 | No | ||
| 69 | EDG5 | 13319 | -0.815 | -0.4756 | No | ||
| 70 | MYO1F | 13510 | -0.873 | -0.4831 | No | ||
| 71 | TBC1D1 | 13673 | -0.921 | -0.4889 | No | ||
| 72 | PRKCB1 | 13692 | -0.927 | -0.4870 | No | ||
| 73 | ROCK1 | 13717 | -0.937 | -0.4853 | No | ||
| 74 | RPA3 | 13877 | -0.986 | -0.4908 | No | ||
| 75 | KCNMB1 | 13891 | -0.990 | -0.4883 | No | ||
| 76 | IFNGR2 | 13937 | -1.006 | -0.4876 | No | ||
| 77 | LRMP | 14085 | -1.059 | -0.4922 | No | ||
| 78 | FANCG | 14208 | -1.104 | -0.4953 | No | ||
| 79 | TRO | 14226 | -1.111 | -0.4927 | No | ||
| 80 | TCL1A | 14395 | -1.165 | -0.4981 | No | ||
| 81 | MUTYH | 14596 | -1.235 | -0.5050 | No | ||
| 82 | BRD3 | 14703 | -1.279 | -0.5067 | No | ||
| 83 | GMDS | 14839 | -1.331 | -0.5097 | No | ||
| 84 | PSCD1 | 14904 | -1.361 | -0.5089 | No | ||
| 85 | HMGCS2 | 14938 | -1.375 | -0.5063 | No | ||
| 86 | LAPTM5 | 14969 | -1.387 | -0.5035 | No | ||
| 87 | POU2F2 | 15003 | -1.400 | -0.5009 | No | ||
| 88 | TAF10 | 15014 | -1.405 | -0.4970 | No | ||
| 89 | PTPN18 | 15205 | -1.486 | -0.5025 | No | ||
| 90 | ATP6AP1 | 15357 | -1.576 | -0.5057 | No | ||
| 91 | LANCL1 | 15664 | -1.732 | -0.5168 | No | ||
| 92 | CRAT | 15851 | -1.849 | -0.5210 | No | ||
| 93 | BCL7A | 16073 | -1.986 | -0.5266 | Yes | ||
| 94 | PPP2R5A | 16096 | -2.000 | -0.5215 | Yes | ||
| 95 | GNG10 | 16163 | -2.070 | -0.5185 | Yes | ||
| 96 | PIK3CG | 16169 | -2.073 | -0.5122 | Yes | ||
| 97 | BASP1 | 16204 | -2.098 | -0.5074 | Yes | ||
| 98 | ATP6V0A2 | 16231 | -2.120 | -0.5021 | Yes | ||
| 99 | BTG2 | 16278 | -2.162 | -0.4977 | Yes | ||
| 100 | DOK2 | 16383 | -2.256 | -0.4962 | Yes | ||
| 101 | DVL1 | 16387 | -2.261 | -0.4892 | Yes | ||
| 102 | BRF1 | 16443 | -2.308 | -0.4848 | Yes | ||
| 103 | AES | 16583 | -2.442 | -0.4846 | Yes | ||
| 104 | RELB | 16614 | -2.472 | -0.4784 | Yes | ||
| 105 | H2AFX | 16660 | -2.521 | -0.4728 | Yes | ||
| 106 | BRD8 | 16771 | -2.646 | -0.4704 | Yes | ||
| 107 | HDAC5 | 16779 | -2.655 | -0.4623 | Yes | ||
| 108 | GMPR | 16837 | -2.722 | -0.4568 | Yes | ||
| 109 | EPS15 | 16908 | -2.818 | -0.4516 | Yes | ||
| 110 | ADA | 17019 | -2.951 | -0.4482 | Yes | ||
| 111 | CSNK1G2 | 17052 | -2.996 | -0.4405 | Yes | ||
| 112 | CUTL1 | 17067 | -3.016 | -0.4317 | Yes | ||
| 113 | ARPC3 | 17144 | -3.138 | -0.4258 | Yes | ||
| 114 | UROS | 17182 | -3.199 | -0.4177 | Yes | ||
| 115 | KIFAP3 | 17288 | -3.367 | -0.4127 | Yes | ||
| 116 | POLD4 | 17388 | -3.565 | -0.4067 | Yes | ||
| 117 | CD53 | 17476 | -3.750 | -0.3996 | Yes | ||
| 118 | HMGCL | 17505 | -3.831 | -0.3889 | Yes | ||
| 119 | CD37 | 17574 | -3.961 | -0.3800 | Yes | ||
| 120 | RGS10 | 17827 | -4.600 | -0.3791 | Yes | ||
| 121 | TOP3B | 17846 | -4.653 | -0.3653 | Yes | ||
| 122 | LTA | 17854 | -4.683 | -0.3508 | Yes | ||
| 123 | IL2RG | 17859 | -4.705 | -0.3361 | Yes | ||
| 124 | RHBDL1 | 18062 | -5.355 | -0.3301 | Yes | ||
| 125 | ATP2A3 | 18074 | -5.389 | -0.3136 | Yes | ||
| 126 | PDK2 | 18108 | -5.558 | -0.2978 | Yes | ||
| 127 | ZFP36L1 | 18112 | -5.581 | -0.2802 | Yes | ||
| 128 | TNFRSF1A | 18237 | -6.230 | -0.2672 | Yes | ||
| 129 | UCP2 | 18250 | -6.341 | -0.2477 | Yes | ||
| 130 | HBP1 | 18344 | -7.095 | -0.2302 | Yes | ||
| 131 | TAZ | 18380 | -7.393 | -0.2087 | Yes | ||
| 132 | STAT2 | 18412 | -7.685 | -0.1860 | Yes | ||
| 133 | MAP4K2 | 18455 | -8.325 | -0.1619 | Yes | ||
| 134 | KCNH2 | 18459 | -8.368 | -0.1355 | Yes | ||
| 135 | CTDSP2 | 18466 | -8.539 | -0.1087 | Yes | ||
| 136 | BCL6 | 18511 | -9.435 | -0.0812 | Yes | ||
| 137 | PTK2B | 18534 | -10.458 | -0.0492 | Yes | ||
| 138 | IL4R | 18586 | -16.906 | 0.0016 | Yes |
