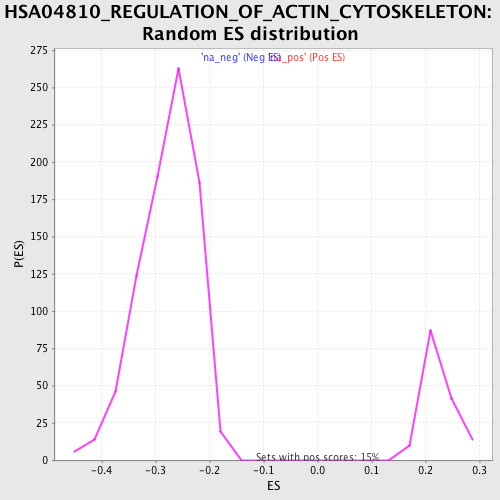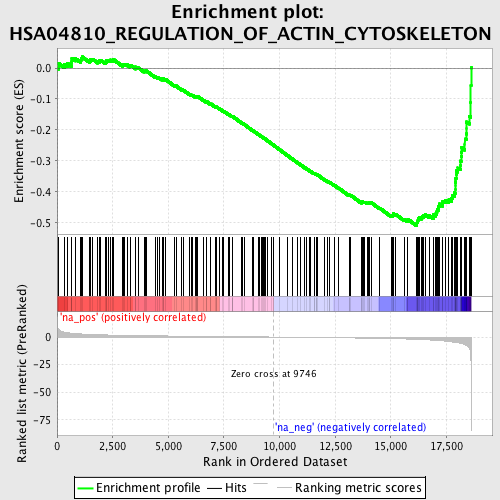
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA04810_REGULATION_OF_ACTIN_CYTOSKELETON |
| Enrichment Score (ES) | -0.51001185 |
| Normalized Enrichment Score (NES) | -1.8394002 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14023507 |
| FWER p-Value | 0.939 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PTK2 | 64 | 7.089 | 0.0148 | No | ||
| 2 | F2 | 323 | 4.441 | 0.0122 | No | ||
| 3 | CFL1 | 458 | 3.955 | 0.0151 | No | ||
| 4 | PIK3R3 | 654 | 3.483 | 0.0135 | No | ||
| 5 | VCL | 661 | 3.467 | 0.0221 | No | ||
| 6 | CYFIP1 | 666 | 3.461 | 0.0308 | No | ||
| 7 | CRK | 816 | 3.207 | 0.0309 | No | ||
| 8 | CD14 | 1049 | 2.871 | 0.0257 | No | ||
| 9 | ITGB5 | 1077 | 2.832 | 0.0316 | No | ||
| 10 | ITGB1 | 1126 | 2.766 | 0.0361 | No | ||
| 11 | PDGFRA | 1473 | 2.373 | 0.0234 | No | ||
| 12 | PIK3R2 | 1505 | 2.342 | 0.0278 | No | ||
| 13 | NCKAP1 | 1595 | 2.272 | 0.0288 | No | ||
| 14 | ACTN3 | 1832 | 2.105 | 0.0214 | No | ||
| 15 | PDGFRB | 1899 | 2.068 | 0.0231 | No | ||
| 16 | FGFR1 | 1948 | 2.038 | 0.0258 | No | ||
| 17 | SSH1 | 2177 | 1.914 | 0.0183 | No | ||
| 18 | PPP1CB | 2216 | 1.891 | 0.0211 | No | ||
| 19 | ARPC1B | 2222 | 1.888 | 0.0257 | No | ||
| 20 | MYH10 | 2317 | 1.841 | 0.0254 | No | ||
| 21 | RDX | 2392 | 1.804 | 0.0260 | No | ||
| 22 | CDC42 | 2501 | 1.755 | 0.0247 | No | ||
| 23 | CFL2 | 2542 | 1.735 | 0.0269 | No | ||
| 24 | MYL9 | 2939 | 1.568 | 0.0095 | No | ||
| 25 | FGFR3 | 2991 | 1.548 | 0.0107 | No | ||
| 26 | PFN4 | 3038 | 1.530 | 0.0122 | No | ||
| 27 | ITGA9 | 3143 | 1.495 | 0.0104 | No | ||
| 28 | ARPC4 | 3276 | 1.445 | 0.0069 | No | ||
| 29 | MAP2K2 | 3298 | 1.440 | 0.0095 | No | ||
| 30 | FGFR2 | 3533 | 1.355 | 0.0003 | No | ||
| 31 | APC | 3540 | 1.354 | 0.0035 | No | ||
| 32 | MYL7 | 3658 | 1.310 | 0.0005 | No | ||
| 33 | EGF | 3908 | 1.234 | -0.0099 | No | ||
| 34 | ARPC5L | 3956 | 1.221 | -0.0093 | No | ||
| 35 | ARHGEF6 | 4012 | 1.202 | -0.0091 | No | ||
| 36 | KRAS | 4401 | 1.100 | -0.0274 | No | ||
| 37 | ITGA4 | 4523 | 1.071 | -0.0312 | No | ||
| 38 | ROCK2 | 4595 | 1.057 | -0.0323 | No | ||
| 39 | FN1 | 4720 | 1.023 | -0.0364 | No | ||
| 40 | NRAS | 4773 | 1.011 | -0.0366 | No | ||
| 41 | FGF9 | 4787 | 1.008 | -0.0347 | No | ||
| 42 | ARPC1A | 4879 | 0.987 | -0.0371 | No | ||
| 43 | FGF5 | 5293 | 0.886 | -0.0572 | No | ||
| 44 | RAF1 | 5344 | 0.875 | -0.0577 | No | ||
| 45 | GSN | 5604 | 0.820 | -0.0696 | No | ||
| 46 | DIAPH3 | 5676 | 0.804 | -0.0714 | No | ||
| 47 | RAC1 | 5946 | 0.749 | -0.0841 | No | ||
| 48 | FGF4 | 6036 | 0.730 | -0.0870 | No | ||
| 49 | ITGB6 | 6082 | 0.722 | -0.0876 | No | ||
| 50 | PIK3R1 | 6233 | 0.693 | -0.0939 | No | ||
| 51 | MRAS | 6255 | 0.687 | -0.0933 | No | ||
| 52 | CHRM2 | 6267 | 0.685 | -0.0922 | No | ||
| 53 | ITGA11 | 6294 | 0.678 | -0.0918 | No | ||
| 54 | GNA12 | 6313 | 0.674 | -0.0911 | No | ||
| 55 | FGF10 | 6597 | 0.616 | -0.1048 | No | ||
| 56 | FGF21 | 6700 | 0.594 | -0.1088 | No | ||
| 57 | PAK3 | 6718 | 0.589 | -0.1082 | No | ||
| 58 | ITGA10 | 6911 | 0.548 | -0.1172 | No | ||
| 59 | FGF3 | 6916 | 0.547 | -0.1160 | No | ||
| 60 | RAC3 | 7120 | 0.508 | -0.1257 | No | ||
| 61 | FGF16 | 7157 | 0.500 | -0.1264 | No | ||
| 62 | ITGA7 | 7163 | 0.498 | -0.1254 | No | ||
| 63 | PAK2 | 7318 | 0.467 | -0.1325 | No | ||
| 64 | TMSB4X | 7444 | 0.444 | -0.1382 | No | ||
| 65 | TIAM2 | 7483 | 0.437 | -0.1391 | No | ||
| 66 | BCAR1 | 7688 | 0.399 | -0.1492 | No | ||
| 67 | MYL2 | 7732 | 0.389 | -0.1505 | No | ||
| 68 | PFN3 | 7864 | 0.361 | -0.1567 | No | ||
| 69 | ARHGEF12 | 7879 | 0.359 | -0.1565 | No | ||
| 70 | FGF8 | 7901 | 0.354 | -0.1567 | No | ||
| 71 | MOS | 8272 | 0.288 | -0.1761 | No | ||
| 72 | ITGA3 | 8321 | 0.280 | -0.1779 | No | ||
| 73 | RRAS | 8432 | 0.258 | -0.1832 | No | ||
| 74 | FGF7 | 8802 | 0.187 | -0.2028 | No | ||
| 75 | FGD1 | 8840 | 0.182 | -0.2043 | No | ||
| 76 | WASF1 | 8842 | 0.182 | -0.2039 | No | ||
| 77 | HRAS | 9039 | 0.147 | -0.2141 | No | ||
| 78 | GNG12 | 9118 | 0.131 | -0.2180 | No | ||
| 79 | ABI2 | 9177 | 0.120 | -0.2209 | No | ||
| 80 | FGF17 | 9220 | 0.113 | -0.2229 | No | ||
| 81 | PXN | 9295 | 0.097 | -0.2266 | No | ||
| 82 | PIK3CB | 9318 | 0.093 | -0.2276 | No | ||
| 83 | MAP2K1 | 9344 | 0.087 | -0.2287 | No | ||
| 84 | ITGAM | 9474 | 0.061 | -0.2355 | No | ||
| 85 | ITGAV | 9627 | 0.025 | -0.2437 | No | ||
| 86 | MYH14 | 9704 | 0.007 | -0.2478 | No | ||
| 87 | FGF23 | 9737 | 0.002 | -0.2496 | No | ||
| 88 | GNA13 | 10003 | -0.059 | -0.2638 | No | ||
| 89 | ITGAX | 10335 | -0.133 | -0.2814 | No | ||
| 90 | SOS1 | 10560 | -0.179 | -0.2931 | No | ||
| 91 | PIP5K3 | 10790 | -0.223 | -0.3049 | No | ||
| 92 | BDKRB1 | 10941 | -0.256 | -0.3124 | No | ||
| 93 | FGFR4 | 11138 | -0.299 | -0.3222 | No | ||
| 94 | FGF14 | 11201 | -0.310 | -0.3248 | No | ||
| 95 | PAK1 | 11346 | -0.340 | -0.3317 | No | ||
| 96 | TIAM1 | 11381 | -0.346 | -0.3327 | No | ||
| 97 | CHRM5 | 11546 | -0.383 | -0.3406 | No | ||
| 98 | MYLK | 11553 | -0.385 | -0.3399 | No | ||
| 99 | FGF2 | 11646 | -0.406 | -0.3439 | No | ||
| 100 | CHRM4 | 11669 | -0.411 | -0.3440 | No | ||
| 101 | PAK7 | 11698 | -0.418 | -0.3445 | No | ||
| 102 | ACTN2 | 12017 | -0.488 | -0.3605 | No | ||
| 103 | CHRM1 | 12158 | -0.520 | -0.3667 | No | ||
| 104 | FGF20 | 12238 | -0.542 | -0.3696 | No | ||
| 105 | SOS2 | 12257 | -0.546 | -0.3692 | No | ||
| 106 | LIMK1 | 12451 | -0.593 | -0.3781 | No | ||
| 107 | ITGB7 | 12652 | -0.640 | -0.3873 | No | ||
| 108 | FGF18 | 13126 | -0.767 | -0.4110 | No | ||
| 109 | FGF11 | 13142 | -0.773 | -0.4098 | No | ||
| 110 | BDKRB2 | 13192 | -0.785 | -0.4104 | No | ||
| 111 | ARHGEF7 | 13668 | -0.920 | -0.4338 | No | ||
| 112 | ARPC2 | 13696 | -0.929 | -0.4329 | No | ||
| 113 | ROCK1 | 13717 | -0.937 | -0.4316 | No | ||
| 114 | ITGA8 | 13787 | -0.959 | -0.4329 | No | ||
| 115 | IQGAP1 | 13833 | -0.974 | -0.4328 | No | ||
| 116 | MAPK3 | 13948 | -1.009 | -0.4364 | No | ||
| 117 | EGFR | 13992 | -1.027 | -0.4361 | No | ||
| 118 | CHRM3 | 14043 | -1.046 | -0.4361 | No | ||
| 119 | PFN1 | 14045 | -1.047 | -0.4335 | No | ||
| 120 | SLC9A1 | 14111 | -1.067 | -0.4342 | No | ||
| 121 | PIK3CA | 14498 | -1.205 | -0.4521 | No | ||
| 122 | FGF1 | 15032 | -1.413 | -0.4773 | No | ||
| 123 | FGF22 | 15069 | -1.430 | -0.4756 | No | ||
| 124 | PAK4 | 15098 | -1.444 | -0.4734 | No | ||
| 125 | ITGA2B | 15118 | -1.450 | -0.4707 | No | ||
| 126 | MAPK1 | 15209 | -1.490 | -0.4718 | No | ||
| 127 | APC2 | 15609 | -1.703 | -0.4890 | No | ||
| 128 | VAV3 | 15745 | -1.781 | -0.4918 | No | ||
| 129 | PDGFB | 15770 | -1.798 | -0.4884 | No | ||
| 130 | PIK3CG | 16169 | -2.073 | -0.5047 | Yes | ||
| 131 | PPP1R12A | 16176 | -2.078 | -0.4997 | Yes | ||
| 132 | ARPC5 | 16190 | -2.087 | -0.4950 | Yes | ||
| 133 | FGF12 | 16218 | -2.109 | -0.4910 | Yes | ||
| 134 | PPP1CC | 16230 | -2.120 | -0.4862 | Yes | ||
| 135 | ARAF | 16268 | -2.156 | -0.4826 | Yes | ||
| 136 | PIK3CD | 16364 | -2.240 | -0.4820 | Yes | ||
| 137 | FGD3 | 16407 | -2.276 | -0.4784 | Yes | ||
| 138 | WASL | 16479 | -2.345 | -0.4763 | Yes | ||
| 139 | CSK | 16538 | -2.400 | -0.4732 | Yes | ||
| 140 | ITGB3 | 16731 | -2.590 | -0.4770 | Yes | ||
| 141 | RRAS2 | 16913 | -2.821 | -0.4795 | Yes | ||
| 142 | PPP1CA | 16921 | -2.831 | -0.4726 | Yes | ||
| 143 | WASF2 | 17028 | -2.961 | -0.4708 | Yes | ||
| 144 | MSN | 17060 | -3.000 | -0.4647 | Yes | ||
| 145 | LIMK2 | 17076 | -3.030 | -0.4578 | Yes | ||
| 146 | ARPC3 | 17144 | -3.138 | -0.4533 | Yes | ||
| 147 | ITGA5 | 17153 | -3.155 | -0.4456 | Yes | ||
| 148 | SSH3 | 17181 | -3.198 | -0.4389 | Yes | ||
| 149 | RAC2 | 17312 | -3.412 | -0.4371 | Yes | ||
| 150 | VAV2 | 17336 | -3.460 | -0.4295 | Yes | ||
| 151 | ITGB2 | 17454 | -3.701 | -0.4263 | Yes | ||
| 152 | BAIAP2 | 17613 | -4.032 | -0.4245 | Yes | ||
| 153 | ITGAE | 17733 | -4.335 | -0.4198 | Yes | ||
| 154 | WAS | 17765 | -4.434 | -0.4101 | Yes | ||
| 155 | PIP5K1C | 17856 | -4.690 | -0.4029 | Yes | ||
| 156 | CRKL | 17893 | -4.821 | -0.3924 | Yes | ||
| 157 | FGF13 | 17897 | -4.828 | -0.3802 | Yes | ||
| 158 | ACTN4 | 17899 | -4.834 | -0.3678 | Yes | ||
| 159 | F2R | 17921 | -4.889 | -0.3564 | Yes | ||
| 160 | ITGAL | 17935 | -4.914 | -0.3444 | Yes | ||
| 161 | VAV1 | 17943 | -4.944 | -0.3321 | Yes | ||
| 162 | ITGB4 | 18017 | -5.184 | -0.3227 | Yes | ||
| 163 | PDGFA | 18133 | -5.648 | -0.3144 | Yes | ||
| 164 | PIK3R5 | 18141 | -5.704 | -0.3001 | Yes | ||
| 165 | MYH9 | 18160 | -5.806 | -0.2862 | Yes | ||
| 166 | MYLPF | 18175 | -5.849 | -0.2719 | Yes | ||
| 167 | RHOA | 18189 | -5.967 | -0.2572 | Yes | ||
| 168 | PFN2 | 18332 | -6.976 | -0.2470 | Yes | ||
| 169 | SSH2 | 18334 | -6.983 | -0.2291 | Yes | ||
| 170 | PIP5K1B | 18388 | -7.474 | -0.2127 | Yes | ||
| 171 | CYFIP2 | 18398 | -7.597 | -0.1937 | Yes | ||
| 172 | MYLC2PL | 18400 | -7.606 | -0.1742 | Yes | ||
| 173 | ARHGEF1 | 18526 | -10.050 | -0.1551 | Yes | ||
| 174 | SCIN | 18596 | -18.657 | -0.1108 | Yes | ||
| 175 | PIP5K1A | 18602 | -20.953 | -0.0572 | Yes | ||
| 176 | ITGA6 | 18607 | -22.511 | 0.0005 | Yes |