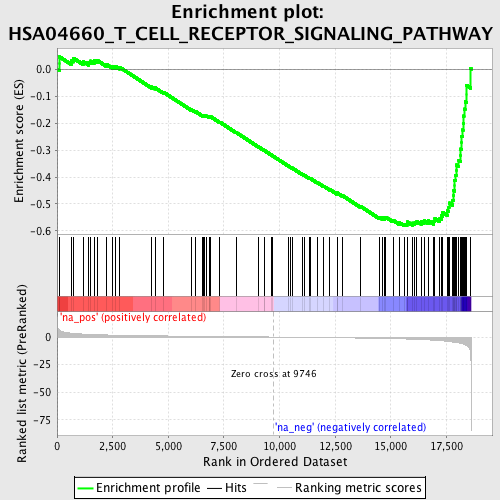
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA04660_T_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.5815193 |
| Normalized Enrichment Score (NES) | -1.9374328 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.13406898 |
| FWER p-Value | 0.729 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL5 | 109 | 6.162 | 0.0198 | No | ||
| 2 | CDK4 | 111 | 6.134 | 0.0454 | No | ||
| 3 | PIK3R3 | 654 | 3.483 | 0.0306 | No | ||
| 4 | GRB2 | 736 | 3.346 | 0.0402 | No | ||
| 5 | NFKBIE | 1172 | 2.703 | 0.0280 | No | ||
| 6 | PTPRC | 1422 | 2.430 | 0.0247 | No | ||
| 7 | PIK3R2 | 1505 | 2.342 | 0.0301 | No | ||
| 8 | PPP3R1 | 1664 | 2.223 | 0.0308 | No | ||
| 9 | NFKBIB | 1814 | 2.114 | 0.0316 | No | ||
| 10 | NCK1 | 2241 | 1.878 | 0.0165 | No | ||
| 11 | CDC42 | 2501 | 1.755 | 0.0098 | No | ||
| 12 | PLCG1 | 2622 | 1.699 | 0.0104 | No | ||
| 13 | PPP3CB | 2813 | 1.620 | 0.0069 | No | ||
| 14 | NFATC4 | 4253 | 1.142 | -0.0660 | No | ||
| 15 | KRAS | 4401 | 1.100 | -0.0693 | No | ||
| 16 | NRAS | 4773 | 1.011 | -0.0851 | No | ||
| 17 | IL4 | 6034 | 0.730 | -0.1501 | No | ||
| 18 | PIK3R1 | 6233 | 0.693 | -0.1579 | No | ||
| 19 | PPP3R2 | 6547 | 0.625 | -0.1722 | No | ||
| 20 | GRAP2 | 6593 | 0.616 | -0.1720 | No | ||
| 21 | NFKB1 | 6645 | 0.606 | -0.1723 | No | ||
| 22 | PAK3 | 6718 | 0.589 | -0.1737 | No | ||
| 23 | IL2 | 6830 | 0.565 | -0.1773 | No | ||
| 24 | CHUK | 6859 | 0.560 | -0.1765 | No | ||
| 25 | AKT1 | 6874 | 0.557 | -0.1749 | No | ||
| 26 | PAK2 | 7318 | 0.467 | -0.1969 | No | ||
| 27 | TNF | 8047 | 0.328 | -0.2348 | No | ||
| 28 | HRAS | 9039 | 0.147 | -0.2877 | No | ||
| 29 | PIK3CB | 9318 | 0.093 | -0.3023 | No | ||
| 30 | CSF2 | 9331 | 0.090 | -0.3026 | No | ||
| 31 | PDCD1 | 9632 | 0.024 | -0.3187 | No | ||
| 32 | CBLC | 9682 | 0.012 | -0.3213 | No | ||
| 33 | MALT1 | 10394 | -0.146 | -0.3590 | No | ||
| 34 | JUN | 10501 | -0.169 | -0.3641 | No | ||
| 35 | SOS1 | 10560 | -0.179 | -0.3665 | No | ||
| 36 | NCK2 | 11043 | -0.278 | -0.3913 | No | ||
| 37 | FOS | 11102 | -0.290 | -0.3932 | No | ||
| 38 | PAK1 | 11346 | -0.340 | -0.4049 | No | ||
| 39 | CD40LG | 11379 | -0.346 | -0.4052 | No | ||
| 40 | PAK7 | 11698 | -0.418 | -0.4206 | No | ||
| 41 | PDK1 | 11983 | -0.481 | -0.4340 | No | ||
| 42 | SOS2 | 12257 | -0.546 | -0.4464 | No | ||
| 43 | NFATC2 | 12580 | -0.623 | -0.4612 | No | ||
| 44 | IFNG | 12581 | -0.623 | -0.4586 | No | ||
| 45 | CBLB | 12833 | -0.686 | -0.4693 | No | ||
| 46 | PPP3CA | 13649 | -0.916 | -0.5094 | No | ||
| 47 | PIK3CA | 14498 | -1.205 | -0.5502 | No | ||
| 48 | PPP3CC | 14611 | -1.241 | -0.5511 | No | ||
| 49 | IKBKB | 14715 | -1.283 | -0.5513 | No | ||
| 50 | IL10 | 14765 | -1.302 | -0.5485 | No | ||
| 51 | PAK4 | 15098 | -1.444 | -0.5604 | No | ||
| 52 | CTLA4 | 15398 | -1.593 | -0.5699 | No | ||
| 53 | ZAP70 | 15615 | -1.707 | -0.5744 | Yes | ||
| 54 | LAT | 15730 | -1.775 | -0.5731 | Yes | ||
| 55 | VAV3 | 15745 | -1.781 | -0.5665 | Yes | ||
| 56 | PTPN6 | 15968 | -1.915 | -0.5705 | Yes | ||
| 57 | AKT2 | 16083 | -1.993 | -0.5683 | Yes | ||
| 58 | PIK3CG | 16169 | -2.073 | -0.5642 | Yes | ||
| 59 | PIK3CD | 16364 | -2.240 | -0.5654 | Yes | ||
| 60 | MAP3K14 | 16495 | -2.360 | -0.5625 | Yes | ||
| 61 | NFKBIA | 16685 | -2.540 | -0.5621 | Yes | ||
| 62 | CD3D | 16927 | -2.841 | -0.5633 | Yes | ||
| 63 | NFKB2 | 16979 | -2.902 | -0.5539 | Yes | ||
| 64 | MAP3K8 | 17204 | -3.241 | -0.5525 | Yes | ||
| 65 | LCK | 17279 | -3.355 | -0.5425 | Yes | ||
| 66 | VAV2 | 17336 | -3.460 | -0.5311 | Yes | ||
| 67 | BCL10 | 17534 | -3.900 | -0.5254 | Yes | ||
| 68 | CBL | 17607 | -4.019 | -0.5126 | Yes | ||
| 69 | RASGRP1 | 17618 | -4.040 | -0.4962 | Yes | ||
| 70 | NFAT5 | 17779 | -4.490 | -0.4861 | Yes | ||
| 71 | CD3G | 17824 | -4.591 | -0.4694 | Yes | ||
| 72 | LCP2 | 17831 | -4.615 | -0.4504 | Yes | ||
| 73 | PRKCQ | 17869 | -4.739 | -0.4326 | Yes | ||
| 74 | NFATC3 | 17871 | -4.741 | -0.4129 | Yes | ||
| 75 | CD4 | 17903 | -4.846 | -0.3944 | Yes | ||
| 76 | IKBKG | 17938 | -4.921 | -0.3757 | Yes | ||
| 77 | VAV1 | 17943 | -4.944 | -0.3553 | Yes | ||
| 78 | FYN | 18056 | -5.334 | -0.3390 | Yes | ||
| 79 | ICOS | 18135 | -5.653 | -0.3197 | Yes | ||
| 80 | PIK3R5 | 18141 | -5.704 | -0.2961 | Yes | ||
| 81 | CARD11 | 18184 | -5.916 | -0.2737 | Yes | ||
| 82 | RHOA | 18189 | -5.967 | -0.2490 | Yes | ||
| 83 | CD247 | 18236 | -6.229 | -0.2255 | Yes | ||
| 84 | CD8B | 18249 | -6.309 | -0.1999 | Yes | ||
| 85 | ITK | 18258 | -6.356 | -0.1738 | Yes | ||
| 86 | CD28 | 18291 | -6.622 | -0.1479 | Yes | ||
| 87 | AKT3 | 18340 | -7.072 | -0.1209 | Yes | ||
| 88 | NFATC1 | 18404 | -7.630 | -0.0925 | Yes | ||
| 89 | CD3E | 18405 | -7.633 | -0.0607 | Yes | ||
| 90 | TEC | 18590 | -17.252 | 0.0014 | Yes |
