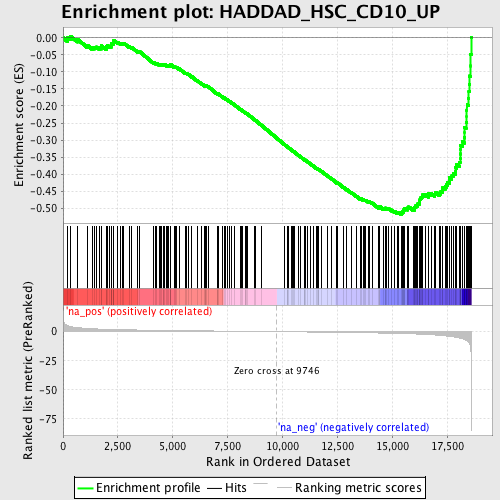
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HADDAD_HSC_CD10_UP |
| Enrichment Score (ES) | -0.518395 |
| Normalized Enrichment Score (NES) | -1.8622168 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.12323199 |
| FWER p-Value | 0.914 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SOD2 | 210 | 5.094 | 0.0008 | No | ||
| 2 | AEBP1 | 330 | 4.414 | 0.0049 | No | ||
| 3 | AKAP12 | 650 | 3.499 | -0.0040 | No | ||
| 4 | RHOB | 1105 | 2.793 | -0.0219 | No | ||
| 5 | COL5A1 | 1328 | 2.516 | -0.0280 | No | ||
| 6 | IL7R | 1421 | 2.430 | -0.0271 | No | ||
| 7 | WFS1 | 1521 | 2.326 | -0.0269 | No | ||
| 8 | SNTB2 | 1670 | 2.219 | -0.0296 | No | ||
| 9 | GNG7 | 1732 | 2.176 | -0.0277 | No | ||
| 10 | QRSL1 | 1746 | 2.165 | -0.0233 | No | ||
| 11 | CTBP2 | 1971 | 2.028 | -0.0306 | No | ||
| 12 | EDEM1 | 1980 | 2.024 | -0.0261 | No | ||
| 13 | ATR | 2012 | 2.005 | -0.0230 | No | ||
| 14 | TIA1 | 2117 | 1.942 | -0.0240 | No | ||
| 15 | FJX1 | 2186 | 1.909 | -0.0231 | No | ||
| 16 | P4HA2 | 2211 | 1.894 | -0.0199 | No | ||
| 17 | GADD45G | 2212 | 1.894 | -0.0154 | No | ||
| 18 | HIST1H3D | 2281 | 1.852 | -0.0146 | No | ||
| 19 | SEC22B | 2282 | 1.852 | -0.0102 | No | ||
| 20 | EIF1AY | 2310 | 1.843 | -0.0072 | No | ||
| 21 | PTPRE | 2488 | 1.759 | -0.0126 | No | ||
| 22 | SNX2 | 2625 | 1.698 | -0.0159 | No | ||
| 23 | HIST1H2AC | 2721 | 1.658 | -0.0171 | No | ||
| 24 | FHIT | 2774 | 1.634 | -0.0160 | No | ||
| 25 | ALDH5A1 | 3013 | 1.540 | -0.0252 | No | ||
| 26 | TOP2A | 3129 | 1.499 | -0.0279 | No | ||
| 27 | ABCG2 | 3404 | 1.396 | -0.0394 | No | ||
| 28 | PHF15 | 3471 | 1.374 | -0.0397 | No | ||
| 29 | CALM1 | 4140 | 1.168 | -0.0732 | No | ||
| 30 | PKD2 | 4214 | 1.150 | -0.0744 | No | ||
| 31 | DNASE2B | 4267 | 1.140 | -0.0744 | No | ||
| 32 | BLNK | 4374 | 1.108 | -0.0775 | No | ||
| 33 | CD24 | 4417 | 1.096 | -0.0772 | No | ||
| 34 | DBN1 | 4503 | 1.077 | -0.0792 | No | ||
| 35 | HIST1H2BG | 4560 | 1.063 | -0.0797 | No | ||
| 36 | CIITA | 4589 | 1.058 | -0.0787 | No | ||
| 37 | VPREB3 | 4633 | 1.049 | -0.0785 | No | ||
| 38 | HIST1H4H | 4719 | 1.024 | -0.0807 | No | ||
| 39 | EFHA1 | 4767 | 1.013 | -0.0808 | No | ||
| 40 | CDKN2C | 4817 | 1.002 | -0.0811 | No | ||
| 41 | TCF3 | 4884 | 0.986 | -0.0823 | No | ||
| 42 | SCHIP1 | 4887 | 0.985 | -0.0800 | No | ||
| 43 | RCBTB1 | 4893 | 0.983 | -0.0780 | No | ||
| 44 | DDR1 | 5075 | 0.941 | -0.0855 | No | ||
| 45 | VDAC1 | 5117 | 0.930 | -0.0855 | No | ||
| 46 | CD9 | 5171 | 0.920 | -0.0862 | No | ||
| 47 | HIST1H3H | 5291 | 0.886 | -0.0905 | No | ||
| 48 | KCNJ2 | 5575 | 0.826 | -0.1039 | No | ||
| 49 | ELL3 | 5610 | 0.819 | -0.1038 | No | ||
| 50 | CAV1 | 5707 | 0.798 | -0.1071 | No | ||
| 51 | PLXNA1 | 5866 | 0.766 | -0.1138 | No | ||
| 52 | ZBTB10 | 6115 | 0.716 | -0.1256 | No | ||
| 53 | CD72 | 6306 | 0.676 | -0.1343 | No | ||
| 54 | CORO2B | 6460 | 0.641 | -0.1410 | No | ||
| 55 | PLEKHF2 | 6481 | 0.637 | -0.1406 | No | ||
| 56 | SH3TC1 | 6493 | 0.635 | -0.1397 | No | ||
| 57 | MME | 6522 | 0.628 | -0.1397 | No | ||
| 58 | HIST1H4B | 6603 | 0.615 | -0.1425 | No | ||
| 59 | CSH2 | 7018 | 0.526 | -0.1637 | No | ||
| 60 | HIST1H2BH | 7058 | 0.519 | -0.1646 | No | ||
| 61 | CD19 | 7072 | 0.516 | -0.1641 | No | ||
| 62 | HIST1H2BF | 7259 | 0.480 | -0.1730 | No | ||
| 63 | NELF | 7337 | 0.464 | -0.1761 | No | ||
| 64 | RIMS3 | 7420 | 0.448 | -0.1795 | No | ||
| 65 | TCF4 | 7482 | 0.437 | -0.1817 | No | ||
| 66 | CACNB3 | 7587 | 0.415 | -0.1864 | No | ||
| 67 | KIF13A | 7691 | 0.398 | -0.1910 | No | ||
| 68 | PCDH9 | 7803 | 0.376 | -0.1961 | No | ||
| 69 | ENTPD4 | 8085 | 0.320 | -0.2106 | No | ||
| 70 | CD79A | 8111 | 0.315 | -0.2112 | No | ||
| 71 | DACT1 | 8177 | 0.303 | -0.2140 | No | ||
| 72 | HIST1H1E | 8305 | 0.282 | -0.2202 | No | ||
| 73 | GPR125 | 8322 | 0.279 | -0.2204 | No | ||
| 74 | MEF2C | 8378 | 0.266 | -0.2228 | No | ||
| 75 | MSX1 | 8381 | 0.266 | -0.2223 | No | ||
| 76 | IL1A | 8713 | 0.203 | -0.2397 | No | ||
| 77 | HMGB2 | 8789 | 0.189 | -0.2434 | No | ||
| 78 | TOP2B | 9026 | 0.149 | -0.2558 | No | ||
| 79 | BANK1 | 10103 | -0.080 | -0.3140 | No | ||
| 80 | ZFYVE16 | 10221 | -0.106 | -0.3201 | No | ||
| 81 | FMR1 | 10231 | -0.107 | -0.3203 | No | ||
| 82 | ARID5B | 10261 | -0.115 | -0.3216 | No | ||
| 83 | GPM6B | 10400 | -0.147 | -0.3288 | No | ||
| 84 | EFNB2 | 10446 | -0.157 | -0.3308 | No | ||
| 85 | JUN | 10501 | -0.169 | -0.3334 | No | ||
| 86 | AK1 | 10536 | -0.176 | -0.3348 | No | ||
| 87 | BLK | 10746 | -0.215 | -0.3456 | No | ||
| 88 | PCBP2 | 10798 | -0.227 | -0.3478 | No | ||
| 89 | NID2 | 11006 | -0.270 | -0.3584 | No | ||
| 90 | PSPC1 | 11017 | -0.273 | -0.3583 | No | ||
| 91 | POU2AF1 | 11033 | -0.276 | -0.3585 | No | ||
| 92 | IGLL1 | 11119 | -0.295 | -0.3624 | No | ||
| 93 | STK38 | 11295 | -0.330 | -0.3711 | No | ||
| 94 | MS4A1 | 11396 | -0.350 | -0.3757 | No | ||
| 95 | MYLK | 11553 | -0.385 | -0.3832 | No | ||
| 96 | POU4F1 | 11611 | -0.397 | -0.3854 | No | ||
| 97 | UXS1 | 11630 | -0.402 | -0.3854 | No | ||
| 98 | NPY | 11656 | -0.409 | -0.3857 | No | ||
| 99 | MAG | 11791 | -0.438 | -0.3920 | No | ||
| 100 | RRM2 | 12069 | -0.498 | -0.4058 | No | ||
| 101 | SCN3A | 12220 | -0.537 | -0.4127 | No | ||
| 102 | HIST1H1C | 12481 | -0.600 | -0.4253 | No | ||
| 103 | SEMA6A | 12520 | -0.609 | -0.4259 | No | ||
| 104 | NGLY1 | 12786 | -0.673 | -0.4387 | No | ||
| 105 | PPM1A | 12935 | -0.712 | -0.4450 | No | ||
| 106 | PCNA | 13131 | -0.768 | -0.4538 | No | ||
| 107 | TPD52 | 13368 | -0.832 | -0.4646 | No | ||
| 108 | RB1CC1 | 13561 | -0.889 | -0.4729 | No | ||
| 109 | STAG3 | 13577 | -0.896 | -0.4716 | No | ||
| 110 | ARHGEF7 | 13668 | -0.920 | -0.4742 | No | ||
| 111 | EFHC1 | 13747 | -0.947 | -0.4762 | No | ||
| 112 | PRDM2 | 13793 | -0.960 | -0.4763 | No | ||
| 113 | ANAPC5 | 13932 | -1.003 | -0.4814 | No | ||
| 114 | P2RY14 | 13958 | -1.014 | -0.4804 | No | ||
| 115 | LRMP | 14085 | -1.059 | -0.4847 | No | ||
| 116 | TCL1A | 14395 | -1.165 | -0.4986 | No | ||
| 117 | RPS11 | 14399 | -1.166 | -0.4960 | No | ||
| 118 | P2RX5 | 14407 | -1.171 | -0.4936 | No | ||
| 119 | PPP3CC | 14611 | -1.241 | -0.5016 | No | ||
| 120 | ITPR1 | 14675 | -1.265 | -0.5020 | No | ||
| 121 | TTC13 | 14682 | -1.269 | -0.4993 | No | ||
| 122 | MEF2A | 14758 | -1.300 | -0.5002 | No | ||
| 123 | PTPRK | 14822 | -1.324 | -0.5005 | No | ||
| 124 | LAPTM5 | 14969 | -1.387 | -0.5051 | No | ||
| 125 | HPS4 | 15119 | -1.452 | -0.5097 | No | ||
| 126 | E2F5 | 15231 | -1.500 | -0.5121 | No | ||
| 127 | COBL | 15284 | -1.530 | -0.5113 | No | ||
| 128 | MAP4K4 | 15416 | -1.601 | -0.5146 | Yes | ||
| 129 | SIAH2 | 15446 | -1.619 | -0.5123 | Yes | ||
| 130 | CXCR4 | 15483 | -1.636 | -0.5103 | Yes | ||
| 131 | RBM6 | 15522 | -1.652 | -0.5084 | Yes | ||
| 132 | CRIM1 | 15531 | -1.657 | -0.5049 | Yes | ||
| 133 | ZHX2 | 15549 | -1.666 | -0.5018 | Yes | ||
| 134 | CITED2 | 15581 | -1.686 | -0.4994 | Yes | ||
| 135 | SLC35D1 | 15693 | -1.744 | -0.5013 | Yes | ||
| 136 | CHD7 | 15716 | -1.768 | -0.4982 | Yes | ||
| 137 | ELF2 | 15759 | -1.789 | -0.4962 | Yes | ||
| 138 | RAD52 | 15965 | -1.915 | -0.5028 | Yes | ||
| 139 | DDX3Y | 16020 | -1.949 | -0.5010 | Yes | ||
| 140 | WSB2 | 16026 | -1.957 | -0.4966 | Yes | ||
| 141 | TOB1 | 16052 | -1.970 | -0.4932 | Yes | ||
| 142 | HS3ST1 | 16113 | -2.016 | -0.4917 | Yes | ||
| 143 | ANKRD10 | 16157 | -2.063 | -0.4891 | Yes | ||
| 144 | AP1B1 | 16172 | -2.074 | -0.4848 | Yes | ||
| 145 | ATM | 16233 | -2.123 | -0.4830 | Yes | ||
| 146 | MDM1 | 16249 | -2.136 | -0.4787 | Yes | ||
| 147 | RB1 | 16263 | -2.150 | -0.4743 | Yes | ||
| 148 | BTG2 | 16278 | -2.162 | -0.4698 | Yes | ||
| 149 | TRIB2 | 16336 | -2.218 | -0.4676 | Yes | ||
| 150 | PIK3CD | 16364 | -2.240 | -0.4637 | Yes | ||
| 151 | LEF1 | 16397 | -2.266 | -0.4600 | Yes | ||
| 152 | IRF4 | 16497 | -2.360 | -0.4597 | Yes | ||
| 153 | JARID1D | 16642 | -2.499 | -0.4616 | Yes | ||
| 154 | MOG | 16648 | -2.503 | -0.4558 | Yes | ||
| 155 | TLE1 | 16795 | -2.674 | -0.4573 | Yes | ||
| 156 | IGF2R | 16943 | -2.855 | -0.4585 | Yes | ||
| 157 | JMJD1C | 16969 | -2.887 | -0.4529 | Yes | ||
| 158 | ING1 | 17151 | -3.151 | -0.4552 | Yes | ||
| 159 | HIP1R | 17217 | -3.254 | -0.4509 | Yes | ||
| 160 | PLXNA2 | 17282 | -3.356 | -0.4464 | Yes | ||
| 161 | PCAF | 17305 | -3.404 | -0.4394 | Yes | ||
| 162 | SOCS2 | 17411 | -3.609 | -0.4364 | Yes | ||
| 163 | NEIL1 | 17485 | -3.791 | -0.4313 | Yes | ||
| 164 | IRF2 | 17520 | -3.862 | -0.4239 | Yes | ||
| 165 | VIL2 | 17602 | -4.005 | -0.4187 | Yes | ||
| 166 | RASGRP1 | 17618 | -4.040 | -0.4098 | Yes | ||
| 167 | EIF2AK3 | 17721 | -4.310 | -0.4051 | Yes | ||
| 168 | FOXO1A | 17778 | -4.489 | -0.3973 | Yes | ||
| 169 | RAG2 | 17872 | -4.745 | -0.3910 | Yes | ||
| 170 | LRIG1 | 17888 | -4.810 | -0.3803 | Yes | ||
| 171 | SMARCA4 | 17941 | -4.934 | -0.3713 | Yes | ||
| 172 | PPM1B | 18059 | -5.342 | -0.3648 | Yes | ||
| 173 | BARD1 | 18096 | -5.490 | -0.3536 | Yes | ||
| 174 | DNTT | 18105 | -5.552 | -0.3408 | Yes | ||
| 175 | ZFP36L1 | 18112 | -5.581 | -0.3277 | Yes | ||
| 176 | CENTD1 | 18119 | -5.603 | -0.3146 | Yes | ||
| 177 | SMAD1 | 18200 | -6.017 | -0.3045 | Yes | ||
| 178 | RAG1 | 18282 | -6.537 | -0.2933 | Yes | ||
| 179 | DCK | 18298 | -6.679 | -0.2781 | Yes | ||
| 180 | CD79B | 18302 | -6.710 | -0.2622 | Yes | ||
| 181 | SSBP2 | 18367 | -7.305 | -0.2481 | Yes | ||
| 182 | CYFIP2 | 18398 | -7.597 | -0.2316 | Yes | ||
| 183 | CD69 | 18401 | -7.609 | -0.2134 | Yes | ||
| 184 | ISG20 | 18408 | -7.656 | -0.1954 | Yes | ||
| 185 | CDKN1B | 18474 | -8.692 | -0.1781 | Yes | ||
| 186 | VPREB1 | 18495 | -9.036 | -0.1576 | Yes | ||
| 187 | VAMP1 | 18512 | -9.484 | -0.1357 | Yes | ||
| 188 | BTG1 | 18531 | -10.306 | -0.1120 | Yes | ||
| 189 | TLE4 | 18572 | -13.147 | -0.0826 | Yes | ||
| 190 | CAPN3 | 18576 | -14.562 | -0.0479 | Yes | ||
| 191 | BACH2 | 18601 | -20.879 | 0.0008 | Yes |
