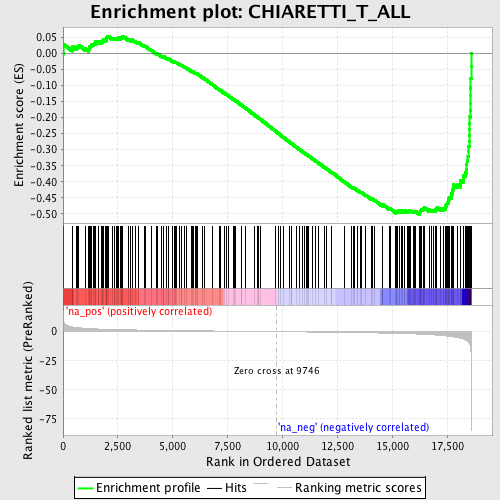
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CHIARETTI_T_ALL |
| Enrichment Score (ES) | -0.50206137 |
| Normalized Enrichment Score (NES) | -1.8383951 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.13398734 |
| FWER p-Value | 0.94 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CEBPB | 31 | 8.104 | 0.0134 | No | ||
| 2 | LGALS1 | 32 | 7.997 | 0.0283 | No | ||
| 3 | BUB1B | 413 | 4.097 | 0.0153 | No | ||
| 4 | PF4 | 437 | 4.014 | 0.0215 | No | ||
| 5 | USP1 | 625 | 3.544 | 0.0180 | No | ||
| 6 | DDIT4 | 659 | 3.472 | 0.0227 | No | ||
| 7 | H1F0 | 723 | 3.371 | 0.0255 | No | ||
| 8 | SPINK2 | 1017 | 2.899 | 0.0150 | No | ||
| 9 | NFKBIE | 1172 | 2.703 | 0.0117 | No | ||
| 10 | IGFBP7 | 1182 | 2.689 | 0.0162 | No | ||
| 11 | PPP1R16B | 1195 | 2.669 | 0.0205 | No | ||
| 12 | GNB2L1 | 1247 | 2.605 | 0.0226 | No | ||
| 13 | CUL1 | 1275 | 2.562 | 0.0259 | No | ||
| 14 | MYC | 1312 | 2.533 | 0.0287 | No | ||
| 15 | IFI30 | 1393 | 2.459 | 0.0289 | No | ||
| 16 | IL7R | 1421 | 2.430 | 0.0320 | No | ||
| 17 | MELK | 1471 | 2.375 | 0.0337 | No | ||
| 18 | LMO2 | 1482 | 2.361 | 0.0376 | No | ||
| 19 | PBX3 | 1611 | 2.259 | 0.0348 | No | ||
| 20 | GCH1 | 1625 | 2.251 | 0.0383 | No | ||
| 21 | SELENBP1 | 1762 | 2.149 | 0.0349 | No | ||
| 22 | G0S2 | 1794 | 2.127 | 0.0372 | No | ||
| 23 | CCL3 | 1813 | 2.114 | 0.0402 | No | ||
| 24 | CTSH | 1821 | 2.110 | 0.0437 | No | ||
| 25 | FGFR1 | 1948 | 2.038 | 0.0407 | No | ||
| 26 | CTBP2 | 1971 | 2.028 | 0.0433 | No | ||
| 27 | NCF4 | 1972 | 2.027 | 0.0470 | No | ||
| 28 | DUSP3 | 1992 | 2.016 | 0.0498 | No | ||
| 29 | TTK | 2008 | 2.006 | 0.0527 | No | ||
| 30 | NFIL3 | 2051 | 1.986 | 0.0541 | No | ||
| 31 | CD44 | 2249 | 1.872 | 0.0469 | No | ||
| 32 | MX1 | 2321 | 1.839 | 0.0465 | No | ||
| 33 | BHLHB2 | 2358 | 1.819 | 0.0479 | No | ||
| 34 | IFRD1 | 2419 | 1.789 | 0.0480 | No | ||
| 35 | PTPRE | 2488 | 1.759 | 0.0476 | No | ||
| 36 | CDC42 | 2501 | 1.755 | 0.0502 | No | ||
| 37 | RNF144 | 2609 | 1.709 | 0.0475 | No | ||
| 38 | HIST1H4C | 2646 | 1.692 | 0.0487 | No | ||
| 39 | ALCAM | 2660 | 1.685 | 0.0512 | No | ||
| 40 | AQP3 | 2707 | 1.664 | 0.0518 | No | ||
| 41 | HIST1H2AC | 2721 | 1.658 | 0.0542 | No | ||
| 42 | ATP9A | 2964 | 1.558 | 0.0439 | No | ||
| 43 | ACSL1 | 3067 | 1.522 | 0.0412 | No | ||
| 44 | ATF3 | 3089 | 1.513 | 0.0429 | No | ||
| 45 | EMP3 | 3142 | 1.496 | 0.0429 | No | ||
| 46 | HNRPDL | 3313 | 1.432 | 0.0363 | No | ||
| 47 | IRF5 | 3423 | 1.390 | 0.0330 | No | ||
| 48 | CD200 | 3448 | 1.381 | 0.0342 | No | ||
| 49 | GAS2 | 3689 | 1.302 | 0.0236 | No | ||
| 50 | SPRY2 | 3748 | 1.284 | 0.0229 | No | ||
| 51 | SERPINE2 | 4019 | 1.198 | 0.0104 | No | ||
| 52 | SWAP70 | 4263 | 1.141 | -0.0006 | No | ||
| 53 | ST3GAL1 | 4313 | 1.125 | -0.0012 | No | ||
| 54 | NKTR | 4494 | 1.079 | -0.0090 | No | ||
| 55 | HIST1H2BJ | 4588 | 1.058 | -0.0120 | No | ||
| 56 | MLC1 | 4590 | 1.058 | -0.0101 | No | ||
| 57 | IL12A | 4718 | 1.024 | -0.0151 | No | ||
| 58 | SCG2 | 4811 | 1.003 | -0.0182 | No | ||
| 59 | CDKN2C | 4817 | 1.002 | -0.0166 | No | ||
| 60 | RGS2 | 4989 | 0.958 | -0.0241 | No | ||
| 61 | IGJ | 5062 | 0.945 | -0.0263 | No | ||
| 62 | GAB2 | 5106 | 0.933 | -0.0269 | No | ||
| 63 | TAF6L | 5153 | 0.923 | -0.0277 | No | ||
| 64 | MTMR6 | 5298 | 0.885 | -0.0338 | No | ||
| 65 | HOXA9 | 5376 | 0.866 | -0.0364 | No | ||
| 66 | LYN | 5405 | 0.860 | -0.0363 | No | ||
| 67 | HIST1H2BL | 5550 | 0.830 | -0.0426 | No | ||
| 68 | SPOCK2 | 5630 | 0.814 | -0.0454 | No | ||
| 69 | GUCY1A3 | 5842 | 0.771 | -0.0554 | No | ||
| 70 | ZBTB16 | 5875 | 0.764 | -0.0557 | No | ||
| 71 | S100A4 | 5927 | 0.753 | -0.0571 | No | ||
| 72 | CXADR | 6039 | 0.730 | -0.0617 | No | ||
| 73 | RSBN1 | 6080 | 0.723 | -0.0626 | No | ||
| 74 | CXCL2 | 6114 | 0.716 | -0.0630 | No | ||
| 75 | ITPR3 | 6364 | 0.662 | -0.0753 | No | ||
| 76 | PTGER4 | 6462 | 0.641 | -0.0794 | No | ||
| 77 | TIPARP | 6808 | 0.568 | -0.0971 | No | ||
| 78 | RUNX3 | 7109 | 0.509 | -0.1124 | No | ||
| 79 | TSHR | 7179 | 0.495 | -0.1152 | No | ||
| 80 | QKI | 7336 | 0.465 | -0.1228 | No | ||
| 81 | ANXA1 | 7453 | 0.442 | -0.1283 | No | ||
| 82 | SPON1 | 7553 | 0.424 | -0.1329 | No | ||
| 83 | ETV5 | 7770 | 0.383 | -0.1439 | No | ||
| 84 | PCDH9 | 7803 | 0.376 | -0.1449 | No | ||
| 85 | KLF10 | 7868 | 0.361 | -0.1477 | No | ||
| 86 | LMO4 | 8110 | 0.315 | -0.1602 | No | ||
| 87 | RGS1 | 8139 | 0.310 | -0.1612 | No | ||
| 88 | PPBP | 8308 | 0.281 | -0.1698 | No | ||
| 89 | SRPK2 | 8729 | 0.199 | -0.1922 | No | ||
| 90 | WASF1 | 8842 | 0.182 | -0.1980 | No | ||
| 91 | PLK2 | 8896 | 0.172 | -0.2005 | No | ||
| 92 | CDC7 | 8909 | 0.171 | -0.2008 | No | ||
| 93 | TUBB2A | 8995 | 0.155 | -0.2052 | No | ||
| 94 | FRMD4B | 9700 | 0.008 | -0.2434 | No | ||
| 95 | FLT3 | 9819 | -0.016 | -0.2498 | No | ||
| 96 | RYBP | 9911 | -0.035 | -0.2546 | No | ||
| 97 | SKAP2 | 10048 | -0.068 | -0.2619 | No | ||
| 98 | CRADD | 10301 | -0.125 | -0.2753 | No | ||
| 99 | PRKCA | 10410 | -0.150 | -0.2809 | No | ||
| 100 | RCBTB2 | 10428 | -0.153 | -0.2816 | No | ||
| 101 | GARNL1 | 10657 | -0.198 | -0.2936 | No | ||
| 102 | FYB | 10768 | -0.220 | -0.2991 | No | ||
| 103 | CASP8 | 10915 | -0.252 | -0.3066 | No | ||
| 104 | NID2 | 11006 | -0.270 | -0.3110 | No | ||
| 105 | FOS | 11102 | -0.290 | -0.3156 | No | ||
| 106 | IGLL1 | 11119 | -0.295 | -0.3159 | No | ||
| 107 | PSPH | 11167 | -0.305 | -0.3179 | No | ||
| 108 | SLC2A3 | 11375 | -0.344 | -0.3285 | No | ||
| 109 | ARF6 | 11489 | -0.371 | -0.3339 | No | ||
| 110 | SLC4A1 | 11652 | -0.407 | -0.3420 | No | ||
| 111 | AHR | 11912 | -0.466 | -0.3552 | No | ||
| 112 | SC4MOL | 11986 | -0.482 | -0.3582 | No | ||
| 113 | ST18 | 12222 | -0.538 | -0.3700 | No | ||
| 114 | SAP30 | 12252 | -0.546 | -0.3706 | No | ||
| 115 | CLEC11A | 12815 | -0.681 | -0.3998 | No | ||
| 116 | JARID2 | 13149 | -0.774 | -0.4164 | No | ||
| 117 | HOXA10 | 13212 | -0.788 | -0.4183 | No | ||
| 118 | PDLIM1 | 13266 | -0.801 | -0.4197 | No | ||
| 119 | CSDA | 13436 | -0.848 | -0.4273 | No | ||
| 120 | RB1CC1 | 13561 | -0.889 | -0.4324 | No | ||
| 121 | FOSB | 13592 | -0.902 | -0.4324 | No | ||
| 122 | PRDM2 | 13793 | -0.960 | -0.4414 | No | ||
| 123 | UBE2E3 | 14036 | -1.043 | -0.4526 | No | ||
| 124 | LRMP | 14085 | -1.059 | -0.4533 | No | ||
| 125 | ATP11B | 14206 | -1.103 | -0.4577 | No | ||
| 126 | CR2 | 14543 | -1.217 | -0.4737 | No | ||
| 127 | MINPP1 | 14547 | -1.220 | -0.4716 | No | ||
| 128 | FHL1 | 14578 | -1.228 | -0.4709 | No | ||
| 129 | HOXA5 | 14854 | -1.340 | -0.4834 | No | ||
| 130 | PLAGL1 | 14924 | -1.368 | -0.4846 | No | ||
| 131 | SFPQ | 15167 | -1.472 | -0.4950 | No | ||
| 132 | DUSP1 | 15206 | -1.487 | -0.4943 | No | ||
| 133 | CRIP1 | 15214 | -1.492 | -0.4919 | No | ||
| 134 | PRDX2 | 15254 | -1.516 | -0.4912 | No | ||
| 135 | BIRC3 | 15345 | -1.560 | -0.4931 | No | ||
| 136 | ADFP | 15346 | -1.561 | -0.4902 | No | ||
| 137 | HPGD | 15431 | -1.610 | -0.4918 | No | ||
| 138 | IDS | 15450 | -1.621 | -0.4897 | No | ||
| 139 | ALDH1A2 | 15539 | -1.660 | -0.4914 | No | ||
| 140 | GABPB2 | 15540 | -1.661 | -0.4883 | No | ||
| 141 | SLC35D1 | 15693 | -1.744 | -0.4933 | No | ||
| 142 | LAT | 15730 | -1.775 | -0.4920 | No | ||
| 143 | HHEX | 15769 | -1.797 | -0.4907 | No | ||
| 144 | RORA | 15836 | -1.839 | -0.4909 | No | ||
| 145 | PMAIP1 | 15949 | -1.907 | -0.4934 | No | ||
| 146 | DDX3Y | 16020 | -1.949 | -0.4936 | No | ||
| 147 | TOB1 | 16052 | -1.970 | -0.4916 | No | ||
| 148 | HES1 | 16246 | -2.134 | -0.4981 | Yes | ||
| 149 | CD2 | 16265 | -2.152 | -0.4951 | Yes | ||
| 150 | PTTG1IP | 16293 | -2.178 | -0.4925 | Yes | ||
| 151 | STAM | 16301 | -2.185 | -0.4888 | Yes | ||
| 152 | TRIB2 | 16336 | -2.218 | -0.4865 | Yes | ||
| 153 | ACTA1 | 16403 | -2.273 | -0.4858 | Yes | ||
| 154 | EPHB6 | 16423 | -2.290 | -0.4826 | Yes | ||
| 155 | STK17B | 16471 | -2.334 | -0.4808 | Yes | ||
| 156 | MAL | 16680 | -2.538 | -0.4874 | Yes | ||
| 157 | CLIC4 | 16791 | -2.670 | -0.4884 | Yes | ||
| 158 | FXYD2 | 16892 | -2.803 | -0.4886 | Yes | ||
| 159 | NOTCH3 | 16958 | -2.876 | -0.4868 | Yes | ||
| 160 | UGCG | 17006 | -2.936 | -0.4838 | Yes | ||
| 161 | DUSP2 | 17039 | -2.987 | -0.4800 | Yes | ||
| 162 | MPO | 17200 | -3.235 | -0.4827 | Yes | ||
| 163 | PCTK2 | 17328 | -3.446 | -0.4832 | Yes | ||
| 164 | SOCS2 | 17411 | -3.609 | -0.4809 | Yes | ||
| 165 | S100A8 | 17418 | -3.617 | -0.4745 | Yes | ||
| 166 | TAP1 | 17472 | -3.735 | -0.4704 | Yes | ||
| 167 | SELL | 17497 | -3.818 | -0.4646 | Yes | ||
| 168 | ITM2A | 17547 | -3.919 | -0.4600 | Yes | ||
| 169 | CCND2 | 17556 | -3.932 | -0.4531 | Yes | ||
| 170 | RASGRP1 | 17618 | -4.040 | -0.4488 | Yes | ||
| 171 | ETS2 | 17687 | -4.238 | -0.4446 | Yes | ||
| 172 | CD97 | 17702 | -4.274 | -0.4374 | Yes | ||
| 173 | SNF1LK | 17729 | -4.328 | -0.4308 | Yes | ||
| 174 | S100A9 | 17759 | -4.412 | -0.4242 | Yes | ||
| 175 | PIM2 | 17790 | -4.508 | -0.4174 | Yes | ||
| 176 | ARHGEF10 | 17794 | -4.514 | -0.4091 | Yes | ||
| 177 | RAP2B | 17980 | -5.047 | -0.4098 | Yes | ||
| 178 | DNTT | 18105 | -5.552 | -0.4062 | Yes | ||
| 179 | ALAS2 | 18120 | -5.603 | -0.3965 | Yes | ||
| 180 | ARID5A | 18230 | -6.204 | -0.3909 | Yes | ||
| 181 | ITK | 18258 | -6.356 | -0.3805 | Yes | ||
| 182 | SLAMF1 | 18330 | -6.970 | -0.3714 | Yes | ||
| 183 | BCL11A | 18381 | -7.407 | -0.3603 | Yes | ||
| 184 | CD69 | 18401 | -7.609 | -0.3471 | Yes | ||
| 185 | ISG20 | 18408 | -7.656 | -0.3332 | Yes | ||
| 186 | ARNTL | 18452 | -8.287 | -0.3201 | Yes | ||
| 187 | CDKN1B | 18474 | -8.692 | -0.3051 | Yes | ||
| 188 | STAT1 | 18476 | -8.708 | -0.2889 | Yes | ||
| 189 | BCL6 | 18511 | -9.435 | -0.2732 | Yes | ||
| 190 | CD52 | 18523 | -9.882 | -0.2554 | Yes | ||
| 191 | ARL4C | 18528 | -10.209 | -0.2366 | Yes | ||
| 192 | RAB6IP1 | 18535 | -10.499 | -0.2173 | Yes | ||
| 193 | DUSP6 | 18542 | -10.788 | -0.1976 | Yes | ||
| 194 | CCL5 | 18555 | -11.313 | -0.1771 | Yes | ||
| 195 | S100A10 | 18566 | -12.247 | -0.1549 | Yes | ||
| 196 | ARMCX2 | 18569 | -12.449 | -0.1318 | Yes | ||
| 197 | TLE4 | 18572 | -13.147 | -0.1074 | Yes | ||
| 198 | IL4R | 18586 | -16.906 | -0.0766 | Yes | ||
| 199 | TES | 18598 | -19.454 | -0.0410 | Yes | ||
| 200 | ITGA6 | 18607 | -22.511 | 0.0005 | Yes |
