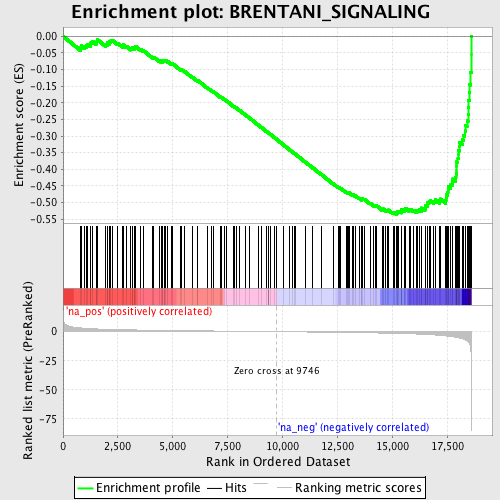
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | BRENTANI_SIGNALING |
| Enrichment Score (ES) | -0.5360499 |
| Normalized Enrichment Score (NES) | -1.9014348 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.124531776 |
| FWER p-Value | 0.833 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAP3K7IP1 | 779 | 3.271 | -0.0341 | No | ||
| 2 | CRK | 816 | 3.207 | -0.0280 | No | ||
| 3 | MAPK10 | 983 | 2.944 | -0.0296 | No | ||
| 4 | ITGB5 | 1077 | 2.832 | -0.0276 | No | ||
| 5 | ITGB1 | 1126 | 2.766 | -0.0233 | No | ||
| 6 | STAT5A | 1251 | 2.601 | -0.0235 | No | ||
| 7 | TNFSF13 | 1268 | 2.574 | -0.0180 | No | ||
| 8 | PTCH2 | 1348 | 2.497 | -0.0160 | No | ||
| 9 | TNFSF4 | 1502 | 2.344 | -0.0184 | No | ||
| 10 | TRAF6 | 1541 | 2.310 | -0.0147 | No | ||
| 11 | JAG2 | 1563 | 2.291 | -0.0101 | No | ||
| 12 | BTK | 1933 | 2.046 | -0.0250 | No | ||
| 13 | TTK | 2008 | 2.006 | -0.0240 | No | ||
| 14 | IL12RB1 | 2019 | 2.002 | -0.0196 | No | ||
| 15 | MAPKAPK2 | 2095 | 1.954 | -0.0188 | No | ||
| 16 | RAN | 2127 | 1.938 | -0.0156 | No | ||
| 17 | TANK | 2180 | 1.913 | -0.0136 | No | ||
| 18 | NCK1 | 2241 | 1.878 | -0.0122 | No | ||
| 19 | MAP2K4 | 2476 | 1.763 | -0.0205 | No | ||
| 20 | TNFRSF1B | 2726 | 1.655 | -0.0298 | No | ||
| 21 | ERG | 2730 | 1.654 | -0.0259 | No | ||
| 22 | IL15 | 2903 | 1.583 | -0.0312 | No | ||
| 23 | YWHAE | 3091 | 1.513 | -0.0376 | No | ||
| 24 | ITGA9 | 3143 | 1.495 | -0.0366 | No | ||
| 25 | TYRO3 | 3165 | 1.485 | -0.0341 | No | ||
| 26 | MAPK13 | 3249 | 1.457 | -0.0349 | No | ||
| 27 | IL1B | 3269 | 1.448 | -0.0323 | No | ||
| 28 | MAP2K2 | 3298 | 1.440 | -0.0303 | No | ||
| 29 | APC | 3540 | 1.354 | -0.0399 | No | ||
| 30 | TNFRSF10B | 3671 | 1.307 | -0.0437 | No | ||
| 31 | WNT4 | 4079 | 1.185 | -0.0628 | No | ||
| 32 | CALM1 | 4140 | 1.168 | -0.0631 | No | ||
| 33 | CXCL1 | 4384 | 1.106 | -0.0736 | No | ||
| 34 | MAPK7 | 4474 | 1.083 | -0.0757 | No | ||
| 35 | ITGA4 | 4523 | 1.071 | -0.0756 | No | ||
| 36 | ARHGAP6 | 4528 | 1.070 | -0.0731 | No | ||
| 37 | CAMK4 | 4601 | 1.056 | -0.0744 | No | ||
| 38 | MAP2K7 | 4635 | 1.048 | -0.0736 | No | ||
| 39 | GRB14 | 4654 | 1.043 | -0.0719 | No | ||
| 40 | NRAS | 4773 | 1.011 | -0.0758 | No | ||
| 41 | SMO | 4926 | 0.975 | -0.0816 | No | ||
| 42 | AXL | 4976 | 0.961 | -0.0819 | No | ||
| 43 | RAF1 | 5344 | 0.875 | -0.0996 | No | ||
| 44 | GRB7 | 5416 | 0.859 | -0.1013 | No | ||
| 45 | IFNB1 | 5531 | 0.833 | -0.1054 | No | ||
| 46 | MCC | 5892 | 0.760 | -0.1230 | No | ||
| 47 | CXCL2 | 6114 | 0.716 | -0.1332 | No | ||
| 48 | NOTCH4 | 6125 | 0.713 | -0.1319 | No | ||
| 49 | ARHGEF16 | 6601 | 0.615 | -0.1562 | No | ||
| 50 | MPL | 6761 | 0.579 | -0.1633 | No | ||
| 51 | AKT1 | 6874 | 0.557 | -0.1680 | No | ||
| 52 | ITGA7 | 7163 | 0.498 | -0.1824 | No | ||
| 53 | RAP1A | 7210 | 0.489 | -0.1836 | No | ||
| 54 | IL13 | 7352 | 0.461 | -0.1901 | No | ||
| 55 | IGFBP3 | 7435 | 0.446 | -0.1935 | No | ||
| 56 | MAP3K4 | 7784 | 0.379 | -0.2114 | No | ||
| 57 | INHA | 7818 | 0.372 | -0.2122 | No | ||
| 58 | ARHGEF12 | 7879 | 0.359 | -0.2146 | No | ||
| 59 | TNF | 8047 | 0.328 | -0.2228 | No | ||
| 60 | ITGA3 | 8321 | 0.280 | -0.2369 | No | ||
| 61 | SKIL | 8501 | 0.244 | -0.2460 | No | ||
| 62 | WNT1 | 8919 | 0.169 | -0.2682 | No | ||
| 63 | HRAS | 9039 | 0.147 | -0.2743 | No | ||
| 64 | ROS1 | 9263 | 0.105 | -0.2861 | No | ||
| 65 | WNT5A | 9343 | 0.088 | -0.2902 | No | ||
| 66 | MAP2K1 | 9344 | 0.087 | -0.2900 | No | ||
| 67 | SUFU | 9352 | 0.085 | -0.2901 | No | ||
| 68 | MAS1 | 9372 | 0.082 | -0.2909 | No | ||
| 69 | ITGAM | 9474 | 0.061 | -0.2963 | No | ||
| 70 | ITGAV | 9627 | 0.025 | -0.3044 | No | ||
| 71 | MAPKAPK5 | 9746 | -0.000 | -0.3108 | No | ||
| 72 | MUSK | 10041 | -0.067 | -0.3266 | No | ||
| 73 | ITGAX | 10335 | -0.133 | -0.3422 | No | ||
| 74 | ABL2 | 10461 | -0.160 | -0.3485 | No | ||
| 75 | SOS1 | 10560 | -0.179 | -0.3534 | No | ||
| 76 | IGFBP1 | 10586 | -0.184 | -0.3543 | No | ||
| 77 | ARHGAP1 | 11045 | -0.278 | -0.3784 | No | ||
| 78 | TIAM1 | 11381 | -0.346 | -0.3957 | No | ||
| 79 | REL | 11773 | -0.433 | -0.4158 | No | ||
| 80 | BIRC2 | 12309 | -0.559 | -0.4434 | No | ||
| 81 | MAPK4 | 12548 | -0.616 | -0.4548 | No | ||
| 82 | IFNG | 12581 | -0.623 | -0.4550 | No | ||
| 83 | ITGB7 | 12652 | -0.640 | -0.4572 | No | ||
| 84 | JAK3 | 12893 | -0.700 | -0.4684 | No | ||
| 85 | BCR | 12982 | -0.724 | -0.4714 | No | ||
| 86 | JAK1 | 13010 | -0.731 | -0.4710 | No | ||
| 87 | DLG1 | 13037 | -0.740 | -0.4706 | No | ||
| 88 | JAG1 | 13173 | -0.779 | -0.4760 | No | ||
| 89 | SRC | 13221 | -0.790 | -0.4765 | No | ||
| 90 | RALBP1 | 13313 | -0.815 | -0.4794 | No | ||
| 91 | MAP3K5 | 13494 | -0.867 | -0.4870 | No | ||
| 92 | TYK2 | 13602 | -0.905 | -0.4906 | No | ||
| 93 | SH3BP2 | 13603 | -0.905 | -0.4883 | No | ||
| 94 | IFNAR1 | 13654 | -0.917 | -0.4887 | No | ||
| 95 | ROCK1 | 13717 | -0.937 | -0.4897 | No | ||
| 96 | WNT2 | 13991 | -1.027 | -0.5020 | No | ||
| 97 | NF1 | 14157 | -1.082 | -0.5082 | No | ||
| 98 | SYK | 14216 | -1.106 | -0.5086 | No | ||
| 99 | MAPK12 | 14274 | -1.127 | -0.5089 | No | ||
| 100 | CXCL12 | 14575 | -1.227 | -0.5221 | No | ||
| 101 | MAP2K3 | 14582 | -1.229 | -0.5193 | No | ||
| 102 | RASA1 | 14704 | -1.279 | -0.5227 | No | ||
| 103 | IL10 | 14765 | -1.302 | -0.5227 | No | ||
| 104 | PTPRK | 14822 | -1.324 | -0.5224 | No | ||
| 105 | MKNK1 | 15044 | -1.418 | -0.5308 | No | ||
| 106 | IRS1 | 15115 | -1.449 | -0.5310 | No | ||
| 107 | MAPK1 | 15209 | -1.490 | -0.5323 | Yes | ||
| 108 | SFN | 15245 | -1.507 | -0.5305 | Yes | ||
| 109 | MYD88 | 15256 | -1.518 | -0.5272 | Yes | ||
| 110 | GNB1 | 15305 | -1.543 | -0.5260 | Yes | ||
| 111 | ABL1 | 15410 | -1.599 | -0.5276 | Yes | ||
| 112 | MAP4K4 | 15416 | -1.601 | -0.5239 | Yes | ||
| 113 | PIK3C3 | 15426 | -1.606 | -0.5204 | Yes | ||
| 114 | MAP2K5 | 15558 | -1.674 | -0.5233 | Yes | ||
| 115 | AXIN2 | 15574 | -1.682 | -0.5199 | Yes | ||
| 116 | ZAP70 | 15615 | -1.707 | -0.5178 | Yes | ||
| 117 | STAT6 | 15767 | -1.794 | -0.5215 | Yes | ||
| 118 | PTEN | 15842 | -1.843 | -0.5209 | Yes | ||
| 119 | STAT5B | 15983 | -1.926 | -0.5237 | Yes | ||
| 120 | AKT2 | 16083 | -1.993 | -0.5241 | Yes | ||
| 121 | PIK3CG | 16169 | -2.073 | -0.5235 | Yes | ||
| 122 | ATM | 16233 | -2.123 | -0.5216 | Yes | ||
| 123 | SHC1 | 16333 | -2.217 | -0.5214 | Yes | ||
| 124 | IFNAR2 | 16345 | -2.223 | -0.5165 | Yes | ||
| 125 | MAP3K14 | 16495 | -2.360 | -0.5187 | Yes | ||
| 126 | ILK | 16520 | -2.381 | -0.5140 | Yes | ||
| 127 | IL2RB | 16526 | -2.388 | -0.5083 | Yes | ||
| 128 | VIPR1 | 16596 | -2.456 | -0.5059 | Yes | ||
| 129 | RELB | 16614 | -2.472 | -0.5007 | Yes | ||
| 130 | IL18R1 | 16677 | -2.537 | -0.4977 | Yes | ||
| 131 | ITGB3 | 16731 | -2.590 | -0.4941 | Yes | ||
| 132 | MAPK9 | 16893 | -2.804 | -0.4958 | Yes | ||
| 133 | NOTCH3 | 16958 | -2.876 | -0.4921 | Yes | ||
| 134 | ITGA5 | 17153 | -3.155 | -0.4947 | Yes | ||
| 135 | MAP3K8 | 17204 | -3.241 | -0.4894 | Yes | ||
| 136 | IFNGR1 | 17448 | -3.681 | -0.4933 | Yes | ||
| 137 | ITGB2 | 17454 | -3.701 | -0.4844 | Yes | ||
| 138 | RELA | 17490 | -3.802 | -0.4768 | Yes | ||
| 139 | BCL10 | 17534 | -3.900 | -0.4694 | Yes | ||
| 140 | MAP2K6 | 17544 | -3.918 | -0.4601 | Yes | ||
| 141 | IL6ST | 17562 | -3.942 | -0.4511 | Yes | ||
| 142 | MAP3K12 | 17675 | -4.196 | -0.4467 | Yes | ||
| 143 | ITGAE | 17733 | -4.335 | -0.4390 | Yes | ||
| 144 | MAPK11 | 17757 | -4.401 | -0.4292 | Yes | ||
| 145 | CRKL | 17893 | -4.821 | -0.4245 | Yes | ||
| 146 | STAT3 | 17907 | -4.850 | -0.4131 | Yes | ||
| 147 | ITGAL | 17935 | -4.914 | -0.4023 | Yes | ||
| 148 | IKBKG | 17938 | -4.921 | -0.3901 | Yes | ||
| 149 | VAV1 | 17943 | -4.944 | -0.3780 | Yes | ||
| 150 | DGKQ | 17996 | -5.108 | -0.3680 | Yes | ||
| 151 | ITGB4 | 18017 | -5.184 | -0.3562 | Yes | ||
| 152 | MAP3K3 | 18023 | -5.201 | -0.3434 | Yes | ||
| 153 | FYN | 18056 | -5.334 | -0.3319 | Yes | ||
| 154 | PDPK1 | 18064 | -5.362 | -0.3188 | Yes | ||
| 155 | AXIN1 | 18194 | -5.994 | -0.3109 | Yes | ||
| 156 | TNFRSF1A | 18237 | -6.230 | -0.2976 | Yes | ||
| 157 | TRAF3 | 18317 | -6.862 | -0.2847 | Yes | ||
| 158 | TNFRSF14 | 18331 | -6.975 | -0.2680 | Yes | ||
| 159 | STAT2 | 18412 | -7.685 | -0.2531 | Yes | ||
| 160 | TRAF1 | 18458 | -8.368 | -0.2347 | Yes | ||
| 161 | STAT1 | 18476 | -8.708 | -0.2139 | Yes | ||
| 162 | RGS16 | 18492 | -8.976 | -0.1922 | Yes | ||
| 163 | DGKA | 18516 | -9.602 | -0.1695 | Yes | ||
| 164 | PTK2B | 18534 | -10.458 | -0.1443 | Yes | ||
| 165 | TNFRSF4 | 18581 | -15.715 | -0.1075 | Yes | ||
| 166 | DGKG | 18605 | -21.261 | -0.0557 | Yes | ||
| 167 | ITGA6 | 18607 | -22.511 | 0.0005 | Yes |
