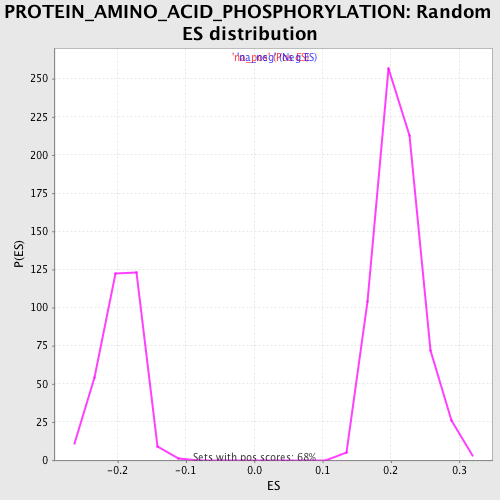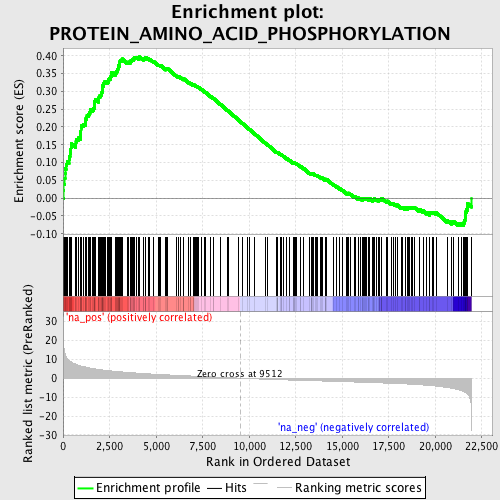
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | PROTEIN_AMINO_ACID_PHOSPHORYLATION |
| Enrichment Score (ES) | 0.39699882 |
| Normalized Enrichment Score (NES) | 1.877116 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05162267 |
| FWER p-Value | 0.58 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LCK | 20 | 19.234 | 0.0211 | Yes | ||
| 2 | SOCS1 | 32 | 17.187 | 0.0403 | Yes | ||
| 3 | CCND3 | 59 | 14.931 | 0.0562 | Yes | ||
| 4 | TEC | 104 | 12.753 | 0.0688 | Yes | ||
| 5 | ABI3 | 112 | 12.643 | 0.0829 | Yes | ||
| 6 | PTK2B | 159 | 11.293 | 0.0938 | Yes | ||
| 7 | MARK2 | 208 | 10.483 | 0.1035 | Yes | ||
| 8 | GMFG | 318 | 9.352 | 0.1092 | Yes | ||
| 9 | TAOK3 | 348 | 9.105 | 0.1183 | Yes | ||
| 10 | MAP4K2 | 372 | 8.936 | 0.1275 | Yes | ||
| 11 | WNK1 | 410 | 8.668 | 0.1357 | Yes | ||
| 12 | CSNK1E | 423 | 8.551 | 0.1450 | Yes | ||
| 13 | ABL1 | 461 | 8.376 | 0.1529 | Yes | ||
| 14 | TGFB1 | 684 | 7.346 | 0.1510 | Yes | ||
| 15 | CLCF1 | 691 | 7.307 | 0.1591 | Yes | ||
| 16 | CSNK2A1 | 727 | 7.148 | 0.1657 | Yes | ||
| 17 | RAF1 | 819 | 6.824 | 0.1693 | Yes | ||
| 18 | CCND2 | 921 | 6.537 | 0.1722 | Yes | ||
| 19 | STK16 | 932 | 6.511 | 0.1792 | Yes | ||
| 20 | MAP3K3 | 939 | 6.496 | 0.1863 | Yes | ||
| 21 | PRKCB1 | 961 | 6.424 | 0.1927 | Yes | ||
| 22 | MAP3K11 | 963 | 6.423 | 0.2000 | Yes | ||
| 23 | MAP4K4 | 1004 | 6.331 | 0.2054 | Yes | ||
| 24 | TXK | 1094 | 6.095 | 0.2083 | Yes | ||
| 25 | MKNK2 | 1193 | 5.865 | 0.2105 | Yes | ||
| 26 | MINK1 | 1214 | 5.832 | 0.2163 | Yes | ||
| 27 | CSK | 1223 | 5.821 | 0.2226 | Yes | ||
| 28 | ULK1 | 1272 | 5.688 | 0.2269 | Yes | ||
| 29 | TLK1 | 1282 | 5.656 | 0.2329 | Yes | ||
| 30 | ACVR1B | 1375 | 5.443 | 0.2349 | Yes | ||
| 31 | TNK2 | 1405 | 5.388 | 0.2398 | Yes | ||
| 32 | IGF1R | 1446 | 5.314 | 0.2440 | Yes | ||
| 33 | ITGB2 | 1471 | 5.262 | 0.2489 | Yes | ||
| 34 | TLK2 | 1596 | 5.060 | 0.2490 | Yes | ||
| 35 | PIM2 | 1646 | 4.980 | 0.2525 | Yes | ||
| 36 | MYLK | 1683 | 4.921 | 0.2564 | Yes | ||
| 37 | CAMK4 | 1688 | 4.913 | 0.2619 | Yes | ||
| 38 | TESK2 | 1692 | 4.905 | 0.2674 | Yes | ||
| 39 | CAMK2B | 1700 | 4.893 | 0.2727 | Yes | ||
| 40 | ALS2CR2 | 1735 | 4.849 | 0.2766 | Yes | ||
| 41 | HIPK3 | 1884 | 4.617 | 0.2751 | Yes | ||
| 42 | CRKRS | 1902 | 4.592 | 0.2796 | Yes | ||
| 43 | COL4A3BP | 1924 | 4.571 | 0.2839 | Yes | ||
| 44 | MAPK3 | 1966 | 4.511 | 0.2871 | Yes | ||
| 45 | DYRK1A | 2032 | 4.420 | 0.2892 | Yes | ||
| 46 | ILK | 2044 | 4.405 | 0.2937 | Yes | ||
| 47 | LMTK2 | 2081 | 4.372 | 0.2971 | Yes | ||
| 48 | TGFBR1 | 2098 | 4.349 | 0.3013 | Yes | ||
| 49 | PINK1 | 2103 | 4.345 | 0.3061 | Yes | ||
| 50 | EIF2AK1 | 2108 | 4.336 | 0.3109 | Yes | ||
| 51 | CSNK1G2 | 2114 | 4.319 | 0.3156 | Yes | ||
| 52 | PDPK1 | 2170 | 4.258 | 0.3180 | Yes | ||
| 53 | IKBKE | 2171 | 4.256 | 0.3228 | Yes | ||
| 54 | MAPK6 | 2196 | 4.227 | 0.3266 | Yes | ||
| 55 | HCLS1 | 2300 | 4.113 | 0.3265 | Yes | ||
| 56 | MAP2K6 | 2378 | 4.033 | 0.3276 | Yes | ||
| 57 | PCTK1 | 2434 | 3.967 | 0.3296 | Yes | ||
| 58 | CSNK1D | 2445 | 3.954 | 0.3337 | Yes | ||
| 59 | ROCK1 | 2484 | 3.908 | 0.3364 | Yes | ||
| 60 | MARK4 | 2537 | 3.861 | 0.3384 | Yes | ||
| 61 | CDC42BPG | 2566 | 3.841 | 0.3416 | Yes | ||
| 62 | DAPK3 | 2588 | 3.821 | 0.3450 | Yes | ||
| 63 | PRKACB | 2592 | 3.816 | 0.3492 | Yes | ||
| 64 | STK38 | 2602 | 3.807 | 0.3531 | Yes | ||
| 65 | CTBP1 | 2795 | 3.610 | 0.3484 | Yes | ||
| 66 | STK4 | 2809 | 3.603 | 0.3520 | Yes | ||
| 67 | LIPE | 2847 | 3.566 | 0.3543 | Yes | ||
| 68 | SRPK2 | 2883 | 3.531 | 0.3568 | Yes | ||
| 69 | PKN2 | 2935 | 3.486 | 0.3584 | Yes | ||
| 70 | STK10 | 2936 | 3.486 | 0.3624 | Yes | ||
| 71 | DYRK2 | 2952 | 3.472 | 0.3657 | Yes | ||
| 72 | DMPK | 2956 | 3.470 | 0.3695 | Yes | ||
| 73 | GSK3B | 2980 | 3.451 | 0.3724 | Yes | ||
| 74 | PCTK2 | 3038 | 3.404 | 0.3737 | Yes | ||
| 75 | STK11 | 3045 | 3.398 | 0.3773 | Yes | ||
| 76 | JAK3 | 3047 | 3.397 | 0.3812 | Yes | ||
| 77 | MAP3K13 | 3051 | 3.396 | 0.3849 | Yes | ||
| 78 | PRKX | 3076 | 3.365 | 0.3877 | Yes | ||
| 79 | CDK9 | 3146 | 3.309 | 0.3883 | Yes | ||
| 80 | MAP4K5 | 3165 | 3.293 | 0.3912 | Yes | ||
| 81 | BRAF | 3459 | 3.052 | 0.3812 | Yes | ||
| 82 | CD81 | 3504 | 3.020 | 0.3826 | Yes | ||
| 83 | ERN1 | 3597 | 2.950 | 0.3818 | Yes | ||
| 84 | NDUFS4 | 3613 | 2.937 | 0.3844 | Yes | ||
| 85 | MAST2 | 3643 | 2.912 | 0.3864 | Yes | ||
| 86 | CAMK2A | 3684 | 2.886 | 0.3879 | Yes | ||
| 87 | PAK1 | 3703 | 2.873 | 0.3904 | Yes | ||
| 88 | RPS6KA4 | 3789 | 2.811 | 0.3897 | Yes | ||
| 89 | AKT1 | 3802 | 2.803 | 0.3923 | Yes | ||
| 90 | STAT1 | 3835 | 2.777 | 0.3940 | Yes | ||
| 91 | STK17B | 3856 | 2.762 | 0.3963 | Yes | ||
| 92 | AKT3 | 3957 | 2.670 | 0.3947 | Yes | ||
| 93 | SGK3 | 4063 | 2.608 | 0.3929 | Yes | ||
| 94 | PKN1 | 4100 | 2.590 | 0.3942 | Yes | ||
| 95 | RAGE | 4104 | 2.584 | 0.3970 | Yes | ||
| 96 | ERC1 | 4304 | 2.438 | 0.3906 | No | ||
| 97 | F2R | 4343 | 2.414 | 0.3916 | No | ||
| 98 | TAF1 | 4411 | 2.376 | 0.3913 | No | ||
| 99 | SNF1LK | 4417 | 2.374 | 0.3938 | No | ||
| 100 | EIF2AK3 | 4441 | 2.359 | 0.3954 | No | ||
| 101 | IL31RA | 4562 | 2.284 | 0.3925 | No | ||
| 102 | DYRK1B | 4666 | 2.220 | 0.3903 | No | ||
| 103 | PXK | 4848 | 2.125 | 0.3844 | No | ||
| 104 | PRKD3 | 5126 | 1.965 | 0.3739 | No | ||
| 105 | MYOD1 | 5171 | 1.940 | 0.3740 | No | ||
| 106 | SNRK | 5253 | 1.898 | 0.3725 | No | ||
| 107 | NEK11 | 5525 | 1.765 | 0.3620 | No | ||
| 108 | PRKG2 | 5536 | 1.761 | 0.3636 | No | ||
| 109 | RPS6KA5 | 5605 | 1.724 | 0.3624 | No | ||
| 110 | IRAK2 | 5619 | 1.715 | 0.3638 | No | ||
| 111 | TYK2 | 6101 | 1.504 | 0.3433 | No | ||
| 112 | JAK2 | 6209 | 1.445 | 0.3401 | No | ||
| 113 | BRSK2 | 6213 | 1.442 | 0.3416 | No | ||
| 114 | PLK3 | 6329 | 1.392 | 0.3379 | No | ||
| 115 | ERBB3 | 6458 | 1.332 | 0.3335 | No | ||
| 116 | BMP4 | 6472 | 1.325 | 0.3344 | No | ||
| 117 | ABI1 | 6483 | 1.320 | 0.3355 | No | ||
| 118 | CDC42BPA | 6748 | 1.195 | 0.3247 | No | ||
| 119 | PICK1 | 6837 | 1.157 | 0.3219 | No | ||
| 120 | PRKCD | 6849 | 1.149 | 0.3227 | No | ||
| 121 | MAP3K2 | 6981 | 1.088 | 0.3180 | No | ||
| 122 | CHUK | 6991 | 1.084 | 0.3188 | No | ||
| 123 | TRIB2 | 7032 | 1.066 | 0.3182 | No | ||
| 124 | ABL2 | 7112 | 1.032 | 0.3157 | No | ||
| 125 | ERBB2 | 7157 | 1.016 | 0.3148 | No | ||
| 126 | AURKA | 7195 | 1.001 | 0.3143 | No | ||
| 127 | PRKCH | 7283 | 0.959 | 0.3114 | No | ||
| 128 | FASTK | 7422 | 0.899 | 0.3060 | No | ||
| 129 | TSSK2 | 7596 | 0.830 | 0.2990 | No | ||
| 130 | CCNT1 | 7634 | 0.813 | 0.2982 | No | ||
| 131 | MERTK | 7941 | 0.688 | 0.2849 | No | ||
| 132 | NUAK2 | 8061 | 0.639 | 0.2802 | No | ||
| 133 | PIM1 | 8068 | 0.635 | 0.2806 | No | ||
| 134 | ROCK2 | 8440 | 0.479 | 0.2641 | No | ||
| 135 | PMVK | 8842 | 0.304 | 0.2459 | No | ||
| 136 | OXSR1 | 8908 | 0.280 | 0.2433 | No | ||
| 137 | PRKAA1 | 9440 | 0.040 | 0.2188 | No | ||
| 138 | EPHA8 | 9624 | -0.056 | 0.2105 | No | ||
| 139 | FYB | 9891 | -0.172 | 0.1984 | No | ||
| 140 | MAP3K8 | 10010 | -0.225 | 0.1932 | No | ||
| 141 | PAK2 | 10297 | -0.335 | 0.1804 | No | ||
| 142 | CARD14 | 10891 | -0.545 | 0.1537 | No | ||
| 143 | MKNK1 | 11007 | -0.582 | 0.1491 | No | ||
| 144 | PLK4 | 11473 | -0.740 | 0.1285 | No | ||
| 145 | DAPK2 | 11495 | -0.747 | 0.1284 | No | ||
| 146 | IL20 | 11498 | -0.748 | 0.1292 | No | ||
| 147 | MAPK4 | 11540 | -0.763 | 0.1281 | No | ||
| 148 | UHMK1 | 11657 | -0.806 | 0.1237 | No | ||
| 149 | MAP4K3 | 11714 | -0.831 | 0.1221 | No | ||
| 150 | TSSK3 | 11719 | -0.832 | 0.1229 | No | ||
| 151 | STK38L | 11856 | -0.876 | 0.1176 | No | ||
| 152 | BTK | 12025 | -0.929 | 0.1109 | No | ||
| 153 | PKN3 | 12153 | -0.961 | 0.1062 | No | ||
| 154 | CAMK1 | 12177 | -0.969 | 0.1062 | No | ||
| 155 | CBLC | 12376 | -1.030 | 0.0983 | No | ||
| 156 | TSSK6 | 12405 | -1.037 | 0.0982 | No | ||
| 157 | CDC42BPB | 12415 | -1.040 | 0.0989 | No | ||
| 158 | CDC2L1 | 12440 | -1.049 | 0.0990 | No | ||
| 159 | RET | 12501 | -1.068 | 0.0975 | No | ||
| 160 | PRKD1 | 12547 | -1.079 | 0.0966 | No | ||
| 161 | ERG | 12732 | -1.135 | 0.0895 | No | ||
| 162 | BCR | 12777 | -1.150 | 0.0888 | No | ||
| 163 | ERN2 | 12928 | -1.194 | 0.0832 | No | ||
| 164 | MAPKAPK2 | 13256 | -1.290 | 0.0696 | No | ||
| 165 | PRKG1 | 13341 | -1.311 | 0.0672 | No | ||
| 166 | MAPK12 | 13362 | -1.319 | 0.0678 | No | ||
| 167 | ADAM10 | 13366 | -1.321 | 0.0692 | No | ||
| 168 | IL3 | 13399 | -1.333 | 0.0693 | No | ||
| 169 | LATS1 | 13438 | -1.344 | 0.0691 | No | ||
| 170 | STK19 | 13583 | -1.388 | 0.0640 | No | ||
| 171 | NEK4 | 13615 | -1.396 | 0.0642 | No | ||
| 172 | PRKCZ | 13684 | -1.423 | 0.0627 | No | ||
| 173 | IRAK3 | 13805 | -1.452 | 0.0588 | No | ||
| 174 | WNK3 | 13879 | -1.473 | 0.0571 | No | ||
| 175 | AURKC | 13921 | -1.484 | 0.0569 | No | ||
| 176 | INSR | 14072 | -1.529 | 0.0518 | No | ||
| 177 | MYO3A | 14090 | -1.533 | 0.0527 | No | ||
| 178 | HCK | 14105 | -1.536 | 0.0539 | No | ||
| 179 | GSG2 | 14156 | -1.552 | 0.0533 | No | ||
| 180 | ABI2 | 14524 | -1.654 | 0.0383 | No | ||
| 181 | IGFBP3 | 14705 | -1.713 | 0.0320 | No | ||
| 182 | CDKL1 | 14860 | -1.757 | 0.0269 | No | ||
| 183 | BMX | 15000 | -1.801 | 0.0226 | No | ||
| 184 | MAP3K10 | 15241 | -1.878 | 0.0136 | No | ||
| 185 | PRKCI | 15299 | -1.894 | 0.0132 | No | ||
| 186 | CAMKK2 | 15318 | -1.900 | 0.0145 | No | ||
| 187 | IL12A | 15437 | -1.935 | 0.0113 | No | ||
| 188 | PTK6 | 15664 | -2.021 | 0.0032 | No | ||
| 189 | BMPR1A | 15701 | -2.033 | 0.0039 | No | ||
| 190 | TSSK4 | 15854 | -2.080 | -0.0007 | No | ||
| 191 | FES | 15874 | -2.085 | 0.0008 | No | ||
| 192 | STK36 | 15953 | -2.109 | -0.0004 | No | ||
| 193 | PRKAG1 | 16085 | -2.147 | -0.0040 | No | ||
| 194 | PRKCE | 16103 | -2.152 | -0.0023 | No | ||
| 195 | WNK2 | 16122 | -2.159 | -0.0007 | No | ||
| 196 | NF2 | 16192 | -2.178 | -0.0014 | No | ||
| 197 | CCND1 | 16273 | -2.210 | -0.0025 | No | ||
| 198 | NEK6 | 16305 | -2.219 | -0.0014 | No | ||
| 199 | EPHA5 | 16406 | -2.252 | -0.0034 | No | ||
| 200 | WNK4 | 16423 | -2.259 | -0.0016 | No | ||
| 201 | EIF2AK4 | 16445 | -2.269 | 0.0001 | No | ||
| 202 | FRK | 16647 | -2.335 | -0.0065 | No | ||
| 203 | ACVR1 | 16677 | -2.349 | -0.0052 | No | ||
| 204 | TNKS | 16690 | -2.352 | -0.0030 | No | ||
| 205 | ICK | 16719 | -2.362 | -0.0016 | No | ||
| 206 | MARK1 | 16819 | -2.403 | -0.0034 | No | ||
| 207 | VRK2 | 16964 | -2.454 | -0.0073 | No | ||
| 208 | LTK | 16979 | -2.459 | -0.0051 | No | ||
| 209 | DAPK1 | 16998 | -2.465 | -0.0031 | No | ||
| 210 | TRIB1 | 17100 | -2.499 | -0.0049 | No | ||
| 211 | MAST1 | 17104 | -2.500 | -0.0022 | No | ||
| 212 | TNK1 | 17115 | -2.504 | 0.0002 | No | ||
| 213 | MATK | 17397 | -2.612 | -0.0097 | No | ||
| 214 | IHPK3 | 17423 | -2.620 | -0.0079 | No | ||
| 215 | SRP72 | 17662 | -2.710 | -0.0157 | No | ||
| 216 | TDGF1 | 17737 | -2.744 | -0.0160 | No | ||
| 217 | CCL11 | 17843 | -2.788 | -0.0176 | No | ||
| 218 | MOBKL1A | 17943 | -2.830 | -0.0190 | No | ||
| 219 | PRPF4B | 18179 | -2.929 | -0.0264 | No | ||
| 220 | EGFR | 18247 | -2.953 | -0.0261 | No | ||
| 221 | GRK1 | 18374 | -3.011 | -0.0285 | No | ||
| 222 | PRKAA2 | 18387 | -3.017 | -0.0256 | No | ||
| 223 | SRPK1 | 18522 | -3.083 | -0.0282 | No | ||
| 224 | MAPK15 | 18544 | -3.092 | -0.0257 | No | ||
| 225 | TTN | 18609 | -3.131 | -0.0250 | No | ||
| 226 | MAP3K9 | 18693 | -3.175 | -0.0252 | No | ||
| 227 | BRSK1 | 18778 | -3.224 | -0.0254 | No | ||
| 228 | STK3 | 18874 | -3.277 | -0.0260 | No | ||
| 229 | CD80 | 19147 | -3.449 | -0.0346 | No | ||
| 230 | MAPK8 | 19156 | -3.453 | -0.0310 | No | ||
| 231 | ACVRL1 | 19338 | -3.567 | -0.0353 | No | ||
| 232 | MAP3K6 | 19547 | -3.725 | -0.0406 | No | ||
| 233 | LATS2 | 19704 | -3.857 | -0.0434 | No | ||
| 234 | LIMK1 | 19712 | -3.863 | -0.0393 | No | ||
| 235 | MAP3K12 | 19825 | -3.968 | -0.0399 | No | ||
| 236 | KALRN | 19924 | -4.058 | -0.0398 | No | ||
| 237 | FGR | 20050 | -4.194 | -0.0407 | No | ||
| 238 | VRK1 | 20630 | -4.884 | -0.0618 | No | ||
| 239 | GMFB | 20872 | -5.285 | -0.0669 | No | ||
| 240 | SNF1LK2 | 20980 | -5.450 | -0.0655 | No | ||
| 241 | CCL2 | 21237 | -6.094 | -0.0704 | No | ||
| 242 | F2 | 21406 | -6.594 | -0.0706 | No | ||
| 243 | IKBKAP | 21537 | -7.074 | -0.0684 | No | ||
| 244 | RIPK4 | 21572 | -7.214 | -0.0617 | No | ||
| 245 | CSNK1A1 | 21599 | -7.335 | -0.0545 | No | ||
| 246 | EPHB2 | 21625 | -7.511 | -0.0471 | No | ||
| 247 | BMPR1B | 21632 | -7.567 | -0.0387 | No | ||
| 248 | CDK2AP1 | 21670 | -7.801 | -0.0315 | No | ||
| 249 | PRDX4 | 21727 | -8.270 | -0.0246 | No | ||
| 250 | DYRK3 | 21738 | -8.370 | -0.0154 | No | ||
| 251 | IL5 | 21945 | -21.856 | 0.0001 | No |