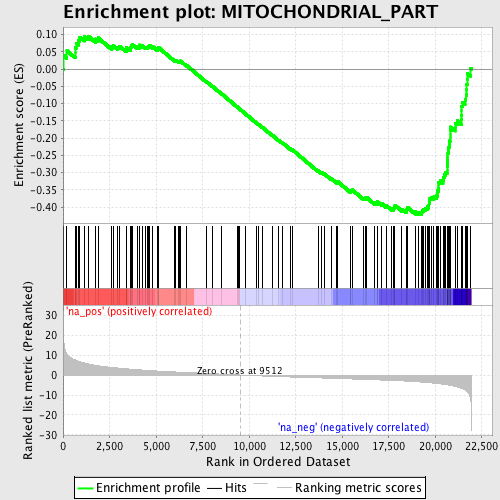
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | -0.42097896 |
| Normalized Enrichment Score (NES) | -1.9838005 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.011449442 |
| FWER p-Value | 0.199 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CASP7 | 14 | 20.405 | 0.0404 | No | ||
| 2 | ALAS2 | 207 | 10.493 | 0.0527 | No | ||
| 3 | SLC25A11 | 653 | 7.471 | 0.0473 | No | ||
| 4 | OXA1L | 655 | 7.462 | 0.0622 | No | ||
| 5 | TIMM17B | 696 | 7.273 | 0.0750 | No | ||
| 6 | RAF1 | 819 | 6.824 | 0.0831 | No | ||
| 7 | MFN2 | 901 | 6.597 | 0.0927 | No | ||
| 8 | BCL2 | 1151 | 5.973 | 0.0933 | No | ||
| 9 | FIS1 | 1347 | 5.512 | 0.0954 | No | ||
| 10 | NDUFA13 | 1742 | 4.842 | 0.0871 | No | ||
| 11 | RHOT2 | 1876 | 4.642 | 0.0903 | No | ||
| 12 | CASQ1 | 2600 | 3.808 | 0.0648 | No | ||
| 13 | NDUFB6 | 2686 | 3.716 | 0.0684 | No | ||
| 14 | MPV17 | 2947 | 3.473 | 0.0634 | No | ||
| 15 | SLC25A22 | 3036 | 3.405 | 0.0663 | No | ||
| 16 | NDUFA2 | 3390 | 3.107 | 0.0563 | No | ||
| 17 | ALDH4A1 | 3396 | 3.105 | 0.0623 | No | ||
| 18 | NDUFS4 | 3613 | 2.937 | 0.0583 | No | ||
| 19 | NFS1 | 3621 | 2.929 | 0.0639 | No | ||
| 20 | OGDH | 3675 | 2.892 | 0.0673 | No | ||
| 21 | ATP5B | 3738 | 2.848 | 0.0702 | No | ||
| 22 | NDUFS8 | 3980 | 2.656 | 0.0645 | No | ||
| 23 | HSD3B2 | 4080 | 2.602 | 0.0651 | No | ||
| 24 | DBT | 4094 | 2.595 | 0.0698 | No | ||
| 25 | NDUFS2 | 4248 | 2.471 | 0.0677 | No | ||
| 26 | MRPL23 | 4443 | 2.358 | 0.0636 | No | ||
| 27 | BCKDK | 4554 | 2.288 | 0.0631 | No | ||
| 28 | ATP5A1 | 4601 | 2.260 | 0.0655 | No | ||
| 29 | SURF1 | 4620 | 2.248 | 0.0692 | No | ||
| 30 | NR3C1 | 4826 | 2.135 | 0.0641 | No | ||
| 31 | COX6B2 | 5064 | 1.993 | 0.0573 | No | ||
| 32 | MCL1 | 5105 | 1.974 | 0.0594 | No | ||
| 33 | HSD3B1 | 5111 | 1.970 | 0.0631 | No | ||
| 34 | NDUFA9 | 5990 | 1.549 | 0.0260 | No | ||
| 35 | NDUFA1 | 6063 | 1.512 | 0.0257 | No | ||
| 36 | TOMM34 | 6214 | 1.442 | 0.0218 | No | ||
| 37 | MRPS21 | 6272 | 1.419 | 0.0220 | No | ||
| 38 | BCKDHB | 6302 | 1.404 | 0.0235 | No | ||
| 39 | MRPS24 | 6646 | 1.233 | 0.0103 | No | ||
| 40 | MRPS16 | 7683 | 0.791 | -0.0356 | No | ||
| 41 | UQCRC1 | 8034 | 0.647 | -0.0504 | No | ||
| 42 | POLG2 | 8507 | 0.449 | -0.0711 | No | ||
| 43 | UCP3 | 9365 | 0.075 | -0.1102 | No | ||
| 44 | BCS1L | 9410 | 0.051 | -0.1122 | No | ||
| 45 | BCKDHA | 9489 | 0.015 | -0.1157 | No | ||
| 46 | MRPL10 | 9818 | -0.141 | -0.1305 | No | ||
| 47 | CENTA2 | 10369 | -0.361 | -0.1549 | No | ||
| 48 | GATM | 10508 | -0.406 | -0.1605 | No | ||
| 49 | ATP5G3 | 10692 | -0.472 | -0.1679 | No | ||
| 50 | NDUFS7 | 11229 | -0.664 | -0.1911 | No | ||
| 51 | ETFB | 11581 | -0.779 | -0.2057 | No | ||
| 52 | ACADM | 11813 | -0.860 | -0.2145 | No | ||
| 53 | TIMM13 | 12200 | -0.975 | -0.2302 | No | ||
| 54 | TFB2M | 12310 | -1.010 | -0.2332 | No | ||
| 55 | MAOB | 13715 | -1.432 | -0.2947 | No | ||
| 56 | ATP5D | 13895 | -1.477 | -0.2999 | No | ||
| 57 | SDHA | 14022 | -1.515 | -0.3027 | No | ||
| 58 | HADHB | 14413 | -1.624 | -0.3173 | No | ||
| 59 | MRPS12 | 14711 | -1.715 | -0.3274 | No | ||
| 60 | UQCRH | 14758 | -1.729 | -0.3261 | No | ||
| 61 | RAB11FIP5 | 15446 | -1.938 | -0.3537 | No | ||
| 62 | ABCB7 | 15458 | -1.942 | -0.3503 | No | ||
| 63 | HCCS | 15536 | -1.975 | -0.3498 | No | ||
| 64 | HTRA2 | 16157 | -2.169 | -0.3739 | No | ||
| 65 | MRPL41 | 16236 | -2.198 | -0.3731 | No | ||
| 66 | NDUFV1 | 16288 | -2.215 | -0.3709 | No | ||
| 67 | ATP5O | 16750 | -2.376 | -0.3873 | No | ||
| 68 | SLC25A1 | 16879 | -2.423 | -0.3883 | No | ||
| 69 | ATP5C1 | 16900 | -2.431 | -0.3843 | No | ||
| 70 | MRPS11 | 17128 | -2.509 | -0.3897 | No | ||
| 71 | GRPEL1 | 17371 | -2.604 | -0.3955 | No | ||
| 72 | NNT | 17651 | -2.705 | -0.4029 | No | ||
| 73 | ATP5G2 | 17774 | -2.762 | -0.4029 | No | ||
| 74 | MSTO1 | 17783 | -2.766 | -0.3977 | No | ||
| 75 | PMPCA | 17832 | -2.786 | -0.3943 | No | ||
| 76 | RHOT1 | 18204 | -2.937 | -0.4055 | No | ||
| 77 | NDUFS3 | 18441 | -3.046 | -0.4101 | No | ||
| 78 | NDUFS1 | 18468 | -3.057 | -0.4052 | No | ||
| 79 | TIMM50 | 18496 | -3.071 | -0.4003 | No | ||
| 80 | UQCRB | 18922 | -3.308 | -0.4131 | No | ||
| 81 | SLC25A15 | 19095 | -3.412 | -0.4141 | Yes | ||
| 82 | MRPS18A | 19244 | -3.505 | -0.4139 | Yes | ||
| 83 | OPA1 | 19287 | -3.529 | -0.4087 | Yes | ||
| 84 | SUPV3L1 | 19378 | -3.595 | -0.4056 | Yes | ||
| 85 | TOMM22 | 19466 | -3.661 | -0.4022 | Yes | ||
| 86 | PPOX | 19558 | -3.733 | -0.3989 | Yes | ||
| 87 | PITRM1 | 19635 | -3.795 | -0.3947 | Yes | ||
| 88 | DNAJA3 | 19660 | -3.816 | -0.3882 | Yes | ||
| 89 | MRPL40 | 19681 | -3.832 | -0.3814 | Yes | ||
| 90 | COX15 | 19709 | -3.862 | -0.3749 | Yes | ||
| 91 | MTX2 | 19794 | -3.941 | -0.3708 | Yes | ||
| 92 | ATP5J | 19927 | -4.060 | -0.3687 | Yes | ||
| 93 | MRPS18C | 20077 | -4.216 | -0.3671 | Yes | ||
| 94 | NDUFAB1 | 20107 | -4.252 | -0.3598 | Yes | ||
| 95 | MRPL32 | 20116 | -4.259 | -0.3516 | Yes | ||
| 96 | MRPS28 | 20172 | -4.313 | -0.3455 | Yes | ||
| 97 | BNIP3 | 20187 | -4.336 | -0.3374 | Yes | ||
| 98 | ABCB6 | 20189 | -4.339 | -0.3287 | Yes | ||
| 99 | ATP5F1 | 20252 | -4.402 | -0.3227 | Yes | ||
| 100 | VDAC3 | 20413 | -4.602 | -0.3208 | Yes | ||
| 101 | MRPL55 | 20435 | -4.641 | -0.3125 | Yes | ||
| 102 | TIMM10 | 20474 | -4.692 | -0.3048 | Yes | ||
| 103 | ACN9 | 20563 | -4.795 | -0.2992 | Yes | ||
| 104 | BAX | 20631 | -4.885 | -0.2924 | Yes | ||
| 105 | CYCS | 20634 | -4.887 | -0.2827 | Yes | ||
| 106 | NDUFA6 | 20649 | -4.914 | -0.2735 | Yes | ||
| 107 | MRPL51 | 20658 | -4.929 | -0.2639 | Yes | ||
| 108 | CS | 20672 | -4.961 | -0.2546 | Yes | ||
| 109 | MRPS35 | 20677 | -4.969 | -0.2447 | Yes | ||
| 110 | PHB2 | 20696 | -5.024 | -0.2355 | Yes | ||
| 111 | ABCB8 | 20716 | -5.059 | -0.2262 | Yes | ||
| 112 | MRPL12 | 20741 | -5.100 | -0.2170 | Yes | ||
| 113 | MRPL52 | 20774 | -5.154 | -0.2081 | Yes | ||
| 114 | MRPS22 | 20792 | -5.186 | -0.1985 | Yes | ||
| 115 | ETFA | 20794 | -5.187 | -0.1881 | Yes | ||
| 116 | ABCF2 | 20796 | -5.189 | -0.1777 | Yes | ||
| 117 | ATP5E | 20805 | -5.200 | -0.1677 | Yes | ||
| 118 | VDAC2 | 21060 | -5.633 | -0.1680 | Yes | ||
| 119 | VDAC1 | 21066 | -5.638 | -0.1569 | Yes | ||
| 120 | TIMM23 | 21174 | -5.898 | -0.1499 | Yes | ||
| 121 | SDHD | 21390 | -6.548 | -0.1466 | Yes | ||
| 122 | TIMM17A | 21399 | -6.579 | -0.1338 | Yes | ||
| 123 | ATP5G1 | 21410 | -6.613 | -0.1209 | Yes | ||
| 124 | TIMM8B | 21413 | -6.619 | -0.1077 | Yes | ||
| 125 | PHB | 21449 | -6.759 | -0.0957 | Yes | ||
| 126 | IMMT | 21596 | -7.320 | -0.0877 | Yes | ||
| 127 | SLC25A3 | 21648 | -7.658 | -0.0747 | Yes | ||
| 128 | TIMM44 | 21690 | -7.999 | -0.0605 | Yes | ||
| 129 | MRPS36 | 21695 | -8.017 | -0.0445 | Yes | ||
| 130 | MRPS15 | 21708 | -8.145 | -0.0287 | Yes | ||
| 131 | TIMM9 | 21713 | -8.175 | -0.0125 | Yes | ||
| 132 | MRPS10 | 21886 | -11.528 | 0.0028 | Yes |
