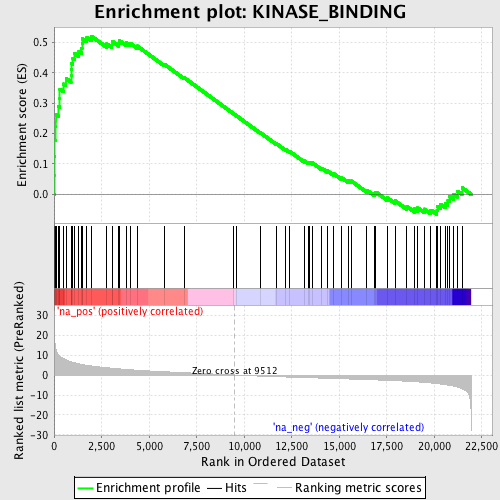
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | KINASE_BINDING |
| Enrichment Score (ES) | 0.52038705 |
| Normalized Enrichment Score (NES) | 2.002777 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.022331502 |
| FWER p-Value | 0.144 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CDKN2D | 15 | 20.092 | 0.0629 | Yes | ||
| 2 | LCK | 20 | 19.234 | 0.1236 | Yes | ||
| 3 | SOCS1 | 32 | 17.187 | 0.1775 | Yes | ||
| 4 | CCND3 | 59 | 14.931 | 0.2236 | Yes | ||
| 5 | SIT1 | 99 | 12.922 | 0.2627 | Yes | ||
| 6 | TOP2B | 230 | 10.225 | 0.2891 | Yes | ||
| 7 | CD4 | 297 | 9.537 | 0.3163 | Yes | ||
| 8 | RAD9A | 304 | 9.493 | 0.3461 | Yes | ||
| 9 | CAMK2N1 | 499 | 8.157 | 0.3630 | Yes | ||
| 10 | TRIP6 | 625 | 7.604 | 0.3814 | Yes | ||
| 11 | KIF13B | 891 | 6.621 | 0.3902 | Yes | ||
| 12 | LAX1 | 898 | 6.608 | 0.4109 | Yes | ||
| 13 | RELA | 916 | 6.549 | 0.4308 | Yes | ||
| 14 | MAP2K7 | 988 | 6.363 | 0.4477 | Yes | ||
| 15 | LLGL1 | 1088 | 6.104 | 0.4625 | Yes | ||
| 16 | CEP250 | 1303 | 5.605 | 0.4705 | Yes | ||
| 17 | IGF1R | 1446 | 5.314 | 0.4808 | Yes | ||
| 18 | ITGB2 | 1471 | 5.262 | 0.4964 | Yes | ||
| 19 | SQSTM1 | 1504 | 5.209 | 0.5114 | Yes | ||
| 20 | FIZ1 | 1724 | 4.863 | 0.5168 | Yes | ||
| 21 | RB1 | 1959 | 4.524 | 0.5204 | Yes | ||
| 22 | BCL10 | 2773 | 3.637 | 0.4947 | No | ||
| 23 | PTPRC | 3046 | 3.398 | 0.4931 | No | ||
| 24 | MAP3K13 | 3051 | 3.396 | 0.5036 | No | ||
| 25 | DLG1 | 3405 | 3.099 | 0.4973 | No | ||
| 26 | MAPK8IP2 | 3456 | 3.057 | 0.5047 | No | ||
| 27 | RPS6KA4 | 3789 | 2.811 | 0.4984 | No | ||
| 28 | APC | 4019 | 2.635 | 0.4963 | No | ||
| 29 | BCAR1 | 4367 | 2.400 | 0.4880 | No | ||
| 30 | PDLIM5 | 5797 | 1.638 | 0.4279 | No | ||
| 31 | PICK1 | 6837 | 1.157 | 0.3841 | No | ||
| 32 | ZC3HC1 | 9436 | 0.041 | 0.2655 | No | ||
| 33 | PTPRR | 9605 | -0.046 | 0.2580 | No | ||
| 34 | AVPR1B | 10851 | -0.530 | 0.2028 | No | ||
| 35 | AKAP11 | 11690 | -0.821 | 0.1671 | No | ||
| 36 | TOP2A | 12184 | -0.971 | 0.1476 | No | ||
| 37 | AKAP7 | 12402 | -1.036 | 0.1410 | No | ||
| 38 | IRS1 | 13166 | -1.265 | 0.1101 | No | ||
| 39 | ADAM10 | 13366 | -1.321 | 0.1052 | No | ||
| 40 | LATS1 | 13438 | -1.344 | 0.1062 | No | ||
| 41 | AKAP5 | 13585 | -1.389 | 0.1039 | No | ||
| 42 | INSR | 14072 | -1.529 | 0.0865 | No | ||
| 43 | TRIB3 | 14369 | -1.612 | 0.0781 | No | ||
| 44 | CDKN2B | 14696 | -1.710 | 0.0686 | No | ||
| 45 | CDKN2C | 15110 | -1.840 | 0.0556 | No | ||
| 46 | UBQLN1 | 15467 | -1.946 | 0.0455 | No | ||
| 47 | CD24 | 15636 | -2.010 | 0.0442 | No | ||
| 48 | AVPR1A | 16455 | -2.272 | 0.0140 | No | ||
| 49 | SPDYA | 16858 | -2.416 | 0.0032 | No | ||
| 50 | AKAP4 | 16929 | -2.443 | 0.0078 | No | ||
| 51 | GRK5 | 17526 | -2.656 | -0.0111 | No | ||
| 52 | MOBKL1A | 17943 | -2.830 | -0.0211 | No | ||
| 53 | FAF1 | 18567 | -3.102 | -0.0398 | No | ||
| 54 | AKAP12 | 18976 | -3.339 | -0.0478 | No | ||
| 55 | CDKN2A | 19143 | -3.446 | -0.0445 | No | ||
| 56 | IRAK1 | 19476 | -3.666 | -0.0481 | No | ||
| 57 | MAP3K12 | 19825 | -3.968 | -0.0514 | No | ||
| 58 | CDK5RAP2 | 20148 | -4.288 | -0.0526 | No | ||
| 59 | MAPK8IP3 | 20178 | -4.329 | -0.0402 | No | ||
| 60 | ADAM9 | 20336 | -4.490 | -0.0331 | No | ||
| 61 | YWHAG | 20590 | -4.829 | -0.0294 | No | ||
| 62 | PRDX3 | 20726 | -5.075 | -0.0195 | No | ||
| 63 | TPRKB | 20823 | -5.221 | -0.0074 | No | ||
| 64 | AKAP3 | 21038 | -5.576 | 0.0005 | No | ||
| 65 | MAPK8IP1 | 21244 | -6.119 | 0.0105 | No | ||
| 66 | HINT1 | 21470 | -6.826 | 0.0218 | No |