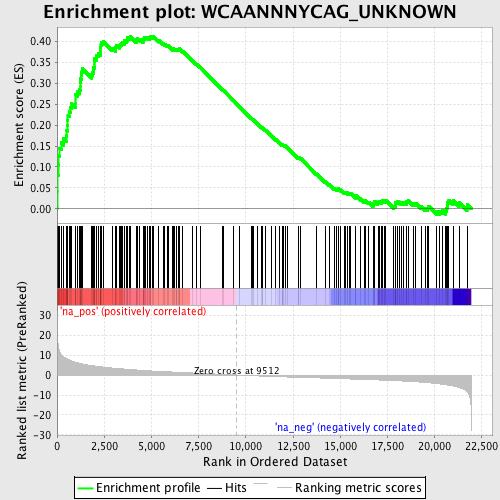
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | WCAANNNYCAG_UNKNOWN |
| Enrichment Score (ES) | 0.41299152 |
| Normalized Enrichment Score (NES) | 1.8533151 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.00908779 |
| FWER p-Value | 0.077 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EGR3 | 3 | 25.122 | 0.0418 | Yes | ||
| 2 | SCN2B | 5 | 23.517 | 0.0810 | Yes | ||
| 3 | BCL2L1 | 52 | 15.577 | 0.1048 | Yes | ||
| 4 | BCL6 | 77 | 13.547 | 0.1263 | Yes | ||
| 5 | ZBTB9 | 118 | 12.444 | 0.1453 | Yes | ||
| 6 | ARF6 | 219 | 10.345 | 0.1579 | Yes | ||
| 7 | PHF12 | 345 | 9.131 | 0.1674 | Yes | ||
| 8 | SERTAD3 | 490 | 8.198 | 0.1745 | Yes | ||
| 9 | PGPEP1 | 512 | 8.087 | 0.1870 | Yes | ||
| 10 | PLCG1 | 539 | 7.963 | 0.1991 | Yes | ||
| 11 | TMEM24 | 550 | 7.916 | 0.2119 | Yes | ||
| 12 | MSI2 | 575 | 7.805 | 0.2238 | Yes | ||
| 13 | SLC4A2 | 665 | 7.427 | 0.2321 | Yes | ||
| 14 | NDUFA3 | 714 | 7.200 | 0.2419 | Yes | ||
| 15 | BANP | 754 | 7.044 | 0.2518 | Yes | ||
| 16 | PLEKHB1 | 986 | 6.365 | 0.2519 | Yes | ||
| 17 | WBP2 | 992 | 6.356 | 0.2622 | Yes | ||
| 18 | JARID2 | 993 | 6.352 | 0.2728 | Yes | ||
| 19 | CNOT4 | 1072 | 6.146 | 0.2795 | Yes | ||
| 20 | CACNB3 | 1181 | 5.905 | 0.2844 | Yes | ||
| 21 | DOCK11 | 1222 | 5.822 | 0.2923 | Yes | ||
| 22 | TUSC4 | 1225 | 5.817 | 0.3019 | Yes | ||
| 23 | KLF7 | 1241 | 5.764 | 0.3108 | Yes | ||
| 24 | CREB1 | 1283 | 5.654 | 0.3184 | Yes | ||
| 25 | TAZ | 1296 | 5.624 | 0.3272 | Yes | ||
| 26 | CBLL1 | 1325 | 5.565 | 0.3352 | Yes | ||
| 27 | MYLIP | 1818 | 4.726 | 0.3205 | Yes | ||
| 28 | YBX2 | 1882 | 4.626 | 0.3253 | Yes | ||
| 29 | DNAJA2 | 1916 | 4.577 | 0.3314 | Yes | ||
| 30 | COL4A3BP | 1924 | 4.571 | 0.3387 | Yes | ||
| 31 | NPHP4 | 1973 | 4.500 | 0.3440 | Yes | ||
| 32 | LMAN2 | 1975 | 4.498 | 0.3515 | Yes | ||
| 33 | NCAM1 | 1985 | 4.481 | 0.3586 | Yes | ||
| 34 | FOXM1 | 2067 | 4.381 | 0.3622 | Yes | ||
| 35 | NUMA1 | 2105 | 4.343 | 0.3677 | Yes | ||
| 36 | CPNE1 | 2187 | 4.234 | 0.3710 | Yes | ||
| 37 | HCLS1 | 2300 | 4.113 | 0.3728 | Yes | ||
| 38 | RDH14 | 2304 | 4.111 | 0.3795 | Yes | ||
| 39 | CNGA1 | 2308 | 4.108 | 0.3862 | Yes | ||
| 40 | ACTR1A | 2324 | 4.087 | 0.3923 | Yes | ||
| 41 | PRCC | 2345 | 4.071 | 0.3982 | Yes | ||
| 42 | JUP | 2455 | 3.944 | 0.3998 | Yes | ||
| 43 | BCOR | 2930 | 3.491 | 0.3838 | Yes | ||
| 44 | BMF | 3084 | 3.355 | 0.3824 | Yes | ||
| 45 | PURA | 3107 | 3.339 | 0.3870 | Yes | ||
| 46 | CDK9 | 3146 | 3.309 | 0.3907 | Yes | ||
| 47 | TOP3A | 3285 | 3.192 | 0.3897 | Yes | ||
| 48 | ORMDL3 | 3337 | 3.153 | 0.3927 | Yes | ||
| 49 | TBX6 | 3402 | 3.103 | 0.3949 | Yes | ||
| 50 | AP1G2 | 3462 | 3.050 | 0.3973 | Yes | ||
| 51 | ARHGEF2 | 3552 | 2.978 | 0.3982 | Yes | ||
| 52 | KLF5 | 3581 | 2.962 | 0.4018 | Yes | ||
| 53 | RPL7 | 3678 | 2.890 | 0.4022 | Yes | ||
| 54 | OSCAR | 3706 | 2.873 | 0.4058 | Yes | ||
| 55 | INVS | 3725 | 2.858 | 0.4097 | Yes | ||
| 56 | PDE6D | 3847 | 2.767 | 0.4088 | Yes | ||
| 57 | ARID5A | 3884 | 2.736 | 0.4117 | Yes | ||
| 58 | CDK5 | 4194 | 2.513 | 0.4017 | Yes | ||
| 59 | BUB1B | 4223 | 2.491 | 0.4046 | Yes | ||
| 60 | MRPL24 | 4260 | 2.464 | 0.4070 | Yes | ||
| 61 | HCFC1R1 | 4387 | 2.393 | 0.4052 | Yes | ||
| 62 | BCKDK | 4554 | 2.288 | 0.4014 | Yes | ||
| 63 | SMCR8 | 4596 | 2.263 | 0.4033 | Yes | ||
| 64 | ATP5A1 | 4601 | 2.260 | 0.4069 | Yes | ||
| 65 | PSMD4 | 4636 | 2.236 | 0.4091 | Yes | ||
| 66 | RDH10 | 4706 | 2.195 | 0.4096 | Yes | ||
| 67 | PANK3 | 4778 | 2.155 | 0.4099 | Yes | ||
| 68 | SLC25A28 | 4888 | 2.103 | 0.4084 | Yes | ||
| 69 | RALBP1 | 4938 | 2.076 | 0.4096 | Yes | ||
| 70 | OTUB1 | 4941 | 2.075 | 0.4130 | Yes | ||
| 71 | VPS45A | 5058 | 1.998 | 0.4110 | No | ||
| 72 | NEXN | 5116 | 1.968 | 0.4117 | No | ||
| 73 | ZNF8 | 5395 | 1.835 | 0.4020 | No | ||
| 74 | KLK1 | 5615 | 1.719 | 0.3948 | No | ||
| 75 | AAK1 | 5713 | 1.679 | 0.3931 | No | ||
| 76 | SMUG1 | 5846 | 1.618 | 0.3898 | No | ||
| 77 | EPS8L2 | 5886 | 1.598 | 0.3906 | No | ||
| 78 | CHM | 6135 | 1.483 | 0.3817 | No | ||
| 79 | GBX2 | 6151 | 1.474 | 0.3835 | No | ||
| 80 | MAP3K7IP2 | 6233 | 1.435 | 0.3822 | No | ||
| 81 | TRIM23 | 6333 | 1.391 | 0.3799 | No | ||
| 82 | NRAS | 6347 | 1.384 | 0.3817 | No | ||
| 83 | RG9MTD2 | 6405 | 1.359 | 0.3813 | No | ||
| 84 | ERBB3 | 6458 | 1.332 | 0.3811 | No | ||
| 85 | BMP4 | 6472 | 1.325 | 0.3827 | No | ||
| 86 | HOXD10 | 6654 | 1.231 | 0.3765 | No | ||
| 87 | ZBTB26 | 7155 | 1.017 | 0.3552 | No | ||
| 88 | PCDH8 | 7402 | 0.908 | 0.3454 | No | ||
| 89 | PRDM1 | 7584 | 0.833 | 0.3385 | No | ||
| 90 | TNNC1 | 8744 | 0.343 | 0.2859 | No | ||
| 91 | BIN3 | 8835 | 0.306 | 0.2823 | No | ||
| 92 | SLC39A5 | 9368 | 0.073 | 0.2580 | No | ||
| 93 | SP8 | 9690 | -0.081 | 0.2433 | No | ||
| 94 | NEUROD6 | 10287 | -0.332 | 0.2165 | No | ||
| 95 | VILL | 10378 | -0.363 | 0.2130 | No | ||
| 96 | STAG2 | 10386 | -0.368 | 0.2133 | No | ||
| 97 | LRRN5 | 10602 | -0.440 | 0.2042 | No | ||
| 98 | SOX2 | 10843 | -0.526 | 0.1940 | No | ||
| 99 | SLC6A9 | 10859 | -0.532 | 0.1942 | No | ||
| 100 | ARTN | 10880 | -0.540 | 0.1942 | No | ||
| 101 | CYB561D2 | 11042 | -0.594 | 0.1878 | No | ||
| 102 | LMX1B | 11371 | -0.705 | 0.1739 | No | ||
| 103 | KCNJ5 | 11587 | -0.780 | 0.1653 | No | ||
| 104 | OTX1 | 11797 | -0.855 | 0.1572 | No | ||
| 105 | SMARCAL1 | 11918 | -0.893 | 0.1532 | No | ||
| 106 | POU4F3 | 11936 | -0.900 | 0.1539 | No | ||
| 107 | UBL3 | 12011 | -0.924 | 0.1520 | No | ||
| 108 | SALL1 | 12077 | -0.942 | 0.1506 | No | ||
| 109 | SOX1 | 12227 | -0.983 | 0.1454 | No | ||
| 110 | FGFR1 | 12767 | -1.146 | 0.1226 | No | ||
| 111 | HNRPM | 12803 | -1.161 | 0.1229 | No | ||
| 112 | PHTF2 | 12893 | -1.185 | 0.1208 | No | ||
| 113 | CLN5 | 13761 | -1.440 | 0.0834 | No | ||
| 114 | NKX2-3 | 14220 | -1.573 | 0.0650 | No | ||
| 115 | AGXT2 | 14439 | -1.630 | 0.0577 | No | ||
| 116 | SV2A | 14689 | -1.708 | 0.0491 | No | ||
| 117 | WNT9A | 14818 | -1.747 | 0.0461 | No | ||
| 118 | DNAJC5B | 14884 | -1.765 | 0.0461 | No | ||
| 119 | FGFR3 | 14904 | -1.772 | 0.0482 | No | ||
| 120 | QTRTD1 | 15030 | -1.808 | 0.0455 | No | ||
| 121 | MAP3K10 | 15241 | -1.878 | 0.0389 | No | ||
| 122 | PCDH10 | 15295 | -1.891 | 0.0397 | No | ||
| 123 | PPP2R2B | 15396 | -1.924 | 0.0383 | No | ||
| 124 | SYT3 | 15512 | -1.964 | 0.0363 | No | ||
| 125 | RNASEH2A | 15562 | -1.986 | 0.0373 | No | ||
| 126 | THPO | 15822 | -2.070 | 0.0289 | No | ||
| 127 | POLK | 15833 | -2.073 | 0.0319 | No | ||
| 128 | HECTD2 | 16080 | -2.146 | 0.0242 | No | ||
| 129 | STARD13 | 16304 | -2.219 | 0.0177 | No | ||
| 130 | SLC31A2 | 16327 | -2.224 | 0.0204 | No | ||
| 131 | FGF10 | 16511 | -2.292 | 0.0158 | No | ||
| 132 | USH1C | 16743 | -2.372 | 0.0091 | No | ||
| 133 | ATP5O | 16750 | -2.376 | 0.0128 | No | ||
| 134 | DMD | 16796 | -2.392 | 0.0147 | No | ||
| 135 | SCNM1 | 16832 | -2.406 | 0.0171 | No | ||
| 136 | FETUB | 17004 | -2.467 | 0.0134 | No | ||
| 137 | GDNF | 17032 | -2.477 | 0.0163 | No | ||
| 138 | NFIX | 17102 | -2.499 | 0.0173 | No | ||
| 139 | LARS | 17200 | -2.534 | 0.0171 | No | ||
| 140 | DPF3 | 17243 | -2.551 | 0.0194 | No | ||
| 141 | RAB30 | 17332 | -2.587 | 0.0197 | No | ||
| 142 | ENPP6 | 17402 | -2.613 | 0.0209 | No | ||
| 143 | SMAD5 | 17846 | -2.789 | 0.0052 | No | ||
| 144 | NUTF2 | 17912 | -2.817 | 0.0069 | No | ||
| 145 | TTR | 17920 | -2.820 | 0.0113 | No | ||
| 146 | FOXP2 | 17925 | -2.821 | 0.0158 | No | ||
| 147 | SFRS2IP | 18015 | -2.861 | 0.0165 | No | ||
| 148 | SCN8A | 18126 | -2.906 | 0.0163 | No | ||
| 149 | WBP5 | 18258 | -2.958 | 0.0152 | No | ||
| 150 | PROS1 | 18372 | -3.010 | 0.0150 | No | ||
| 151 | TCEA2 | 18499 | -3.072 | 0.0144 | No | ||
| 152 | SUFU | 18528 | -3.087 | 0.0182 | No | ||
| 153 | MYH6 | 18598 | -3.123 | 0.0203 | No | ||
| 154 | SYNCRIP | 18869 | -3.273 | 0.0133 | No | ||
| 155 | CNTFR | 19002 | -3.356 | 0.0129 | No | ||
| 156 | MAPK10 | 19290 | -3.532 | 0.0056 | No | ||
| 157 | ANGPT4 | 19512 | -3.698 | 0.0016 | No | ||
| 158 | GRWD1 | 19651 | -3.808 | 0.0016 | No | ||
| 159 | BTRC | 19694 | -3.850 | 0.0061 | No | ||
| 160 | RG9MTD3 | 20114 | -4.258 | -0.0060 | No | ||
| 161 | TNNI1 | 20281 | -4.430 | -0.0062 | No | ||
| 162 | NMT1 | 20406 | -4.591 | -0.0043 | No | ||
| 163 | YWHAG | 20590 | -4.829 | -0.0046 | No | ||
| 164 | LRP1 | 20637 | -4.891 | 0.0014 | No | ||
| 165 | PRG1 | 20666 | -4.942 | 0.0084 | No | ||
| 166 | CCDC5 | 20680 | -4.975 | 0.0161 | No | ||
| 167 | PPHLN1 | 20761 | -5.129 | 0.0210 | No | ||
| 168 | FBXO36 | 20983 | -5.456 | 0.0199 | No | ||
| 169 | RPP40 | 21300 | -6.292 | 0.0159 | No | ||
| 170 | PA2G4 | 21722 | -8.235 | 0.0103 | No |
