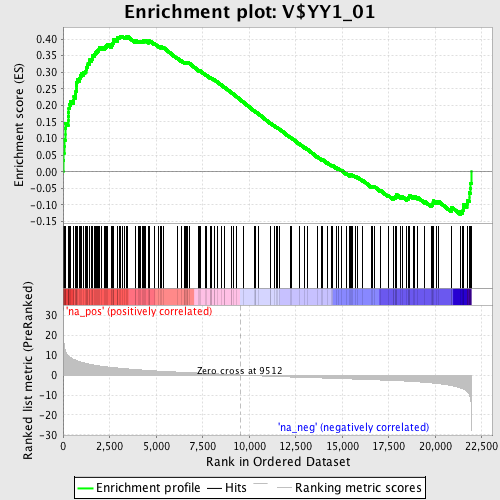
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$YY1_01 |
| Enrichment Score (ES) | 0.40870398 |
| Normalized Enrichment Score (NES) | 1.8432524 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.008919941 |
| FWER p-Value | 0.082 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EGR3 | 3 | 25.122 | 0.0347 | Yes | ||
| 2 | EGR2 | 41 | 16.374 | 0.0557 | Yes | ||
| 3 | CD3E | 51 | 15.588 | 0.0769 | Yes | ||
| 4 | PHF8 | 57 | 15.076 | 0.0975 | Yes | ||
| 5 | CBX4 | 109 | 12.686 | 0.1128 | Yes | ||
| 6 | SCML4 | 111 | 12.656 | 0.1303 | Yes | ||
| 7 | IHPK2 | 131 | 12.123 | 0.1462 | Yes | ||
| 8 | RBMX | 267 | 9.771 | 0.1536 | Yes | ||
| 9 | GBF1 | 273 | 9.723 | 0.1668 | Yes | ||
| 10 | CD4 | 297 | 9.537 | 0.1790 | Yes | ||
| 11 | TRAF3 | 312 | 9.412 | 0.1914 | Yes | ||
| 12 | PHF12 | 345 | 9.131 | 0.2026 | Yes | ||
| 13 | YWHAE | 396 | 8.835 | 0.2125 | Yes | ||
| 14 | RNF44 | 564 | 7.862 | 0.2157 | Yes | ||
| 15 | PRKACA | 580 | 7.756 | 0.2258 | Yes | ||
| 16 | CTDSP1 | 666 | 7.427 | 0.2322 | Yes | ||
| 17 | SRF | 686 | 7.344 | 0.2415 | Yes | ||
| 18 | PRDM10 | 706 | 7.234 | 0.2506 | Yes | ||
| 19 | FALZ | 711 | 7.210 | 0.2605 | Yes | ||
| 20 | PAFAH1B1 | 726 | 7.153 | 0.2697 | Yes | ||
| 21 | RAB1B | 751 | 7.053 | 0.2784 | Yes | ||
| 22 | PFN2 | 890 | 6.623 | 0.2812 | Yes | ||
| 23 | RELA | 916 | 6.549 | 0.2892 | Yes | ||
| 24 | TCTA | 969 | 6.410 | 0.2957 | Yes | ||
| 25 | ASH1L | 1084 | 6.122 | 0.2989 | Yes | ||
| 26 | PPP2R5C | 1176 | 5.914 | 0.3029 | Yes | ||
| 27 | ING4 | 1230 | 5.786 | 0.3085 | Yes | ||
| 28 | ACTG2 | 1264 | 5.712 | 0.3149 | Yes | ||
| 29 | TAZ | 1296 | 5.624 | 0.3213 | Yes | ||
| 30 | PLAG1 | 1336 | 5.534 | 0.3272 | Yes | ||
| 31 | LCP2 | 1403 | 5.391 | 0.3316 | Yes | ||
| 32 | MLL5 | 1438 | 5.327 | 0.3374 | Yes | ||
| 33 | GNAI2 | 1546 | 5.136 | 0.3396 | Yes | ||
| 34 | FBXW11 | 1563 | 5.105 | 0.3460 | Yes | ||
| 35 | TLK2 | 1596 | 5.060 | 0.3515 | Yes | ||
| 36 | RAB1A | 1676 | 4.929 | 0.3547 | Yes | ||
| 37 | TLE3 | 1727 | 4.860 | 0.3592 | Yes | ||
| 38 | RAB22A | 1781 | 4.784 | 0.3634 | Yes | ||
| 39 | PHF6 | 1864 | 4.663 | 0.3661 | Yes | ||
| 40 | ROD1 | 1922 | 4.572 | 0.3698 | Yes | ||
| 41 | UBE4B | 1934 | 4.560 | 0.3756 | Yes | ||
| 42 | KIT | 2072 | 4.379 | 0.3754 | Yes | ||
| 43 | FUS | 2229 | 4.200 | 0.3740 | Yes | ||
| 44 | RGS12 | 2295 | 4.117 | 0.3767 | Yes | ||
| 45 | ZFP91 | 2331 | 4.083 | 0.3808 | Yes | ||
| 46 | KIF1B | 2390 | 4.023 | 0.3837 | Yes | ||
| 47 | CHD2 | 2595 | 3.816 | 0.3796 | Yes | ||
| 48 | CASQ1 | 2600 | 3.808 | 0.3847 | Yes | ||
| 49 | ARMCX3 | 2672 | 3.731 | 0.3866 | Yes | ||
| 50 | DNAJB4 | 2693 | 3.709 | 0.3909 | Yes | ||
| 51 | LUC7L2 | 2721 | 3.683 | 0.3947 | Yes | ||
| 52 | EPC1 | 2723 | 3.682 | 0.3998 | Yes | ||
| 53 | RBMS1 | 2906 | 3.511 | 0.3963 | Yes | ||
| 54 | ANKRD17 | 2918 | 3.502 | 0.4006 | Yes | ||
| 55 | DDX6 | 2925 | 3.495 | 0.4052 | Yes | ||
| 56 | PCF11 | 3029 | 3.410 | 0.4052 | Yes | ||
| 57 | NR2C2 | 3056 | 3.391 | 0.4087 | Yes | ||
| 58 | MAP4K5 | 3165 | 3.293 | 0.4083 | No | ||
| 59 | SNX13 | 3321 | 3.165 | 0.4056 | No | ||
| 60 | AKAP8L | 3399 | 3.104 | 0.4063 | No | ||
| 61 | GTF3C2 | 3452 | 3.062 | 0.4082 | No | ||
| 62 | ATP6V0B | 3887 | 2.734 | 0.3920 | No | ||
| 63 | WTAP | 3893 | 2.730 | 0.3956 | No | ||
| 64 | CTCF | 4032 | 2.627 | 0.3929 | No | ||
| 65 | EML4 | 4122 | 2.573 | 0.3924 | No | ||
| 66 | SRGAP2 | 4176 | 2.530 | 0.3935 | No | ||
| 67 | UBE3A | 4251 | 2.469 | 0.3935 | No | ||
| 68 | FBXW7 | 4301 | 2.440 | 0.3946 | No | ||
| 69 | SRRM2 | 4384 | 2.394 | 0.3942 | No | ||
| 70 | NFE2L1 | 4421 | 2.371 | 0.3958 | No | ||
| 71 | NGFRAP1 | 4597 | 2.263 | 0.3909 | No | ||
| 72 | HIC2 | 4633 | 2.238 | 0.3924 | No | ||
| 73 | OPRM1 | 4659 | 2.223 | 0.3943 | No | ||
| 74 | SSH2 | 4916 | 2.088 | 0.3855 | No | ||
| 75 | PRKAB2 | 5130 | 1.963 | 0.3784 | No | ||
| 76 | DPAGT1 | 5255 | 1.897 | 0.3753 | No | ||
| 77 | GRIN2B | 5286 | 1.882 | 0.3766 | No | ||
| 78 | THRAP4 | 5383 | 1.841 | 0.3747 | No | ||
| 79 | CHM | 6135 | 1.483 | 0.3422 | No | ||
| 80 | NRAS | 6347 | 1.384 | 0.3345 | No | ||
| 81 | NCDN | 6518 | 1.301 | 0.3285 | No | ||
| 82 | CREM | 6538 | 1.289 | 0.3294 | No | ||
| 83 | CAST | 6585 | 1.267 | 0.3290 | No | ||
| 84 | HOXD10 | 6654 | 1.231 | 0.3276 | No | ||
| 85 | CASQ2 | 6661 | 1.228 | 0.3290 | No | ||
| 86 | HOXC10 | 6671 | 1.224 | 0.3303 | No | ||
| 87 | FOS | 6777 | 1.183 | 0.3271 | No | ||
| 88 | CDH6 | 7298 | 0.951 | 0.3046 | No | ||
| 89 | FHL3 | 7333 | 0.936 | 0.3043 | No | ||
| 90 | BNC2 | 7370 | 0.923 | 0.3039 | No | ||
| 91 | PRM1 | 7643 | 0.810 | 0.2926 | No | ||
| 92 | PLXNB1 | 7696 | 0.785 | 0.2912 | No | ||
| 93 | PDZRN4 | 7902 | 0.702 | 0.2828 | No | ||
| 94 | ADNP | 7904 | 0.701 | 0.2837 | No | ||
| 95 | NETO1 | 7977 | 0.672 | 0.2814 | No | ||
| 96 | RHOA | 7991 | 0.666 | 0.2817 | No | ||
| 97 | MYH11 | 8116 | 0.612 | 0.2768 | No | ||
| 98 | POPDC2 | 8127 | 0.606 | 0.2772 | No | ||
| 99 | ECEL1 | 8268 | 0.551 | 0.2715 | No | ||
| 100 | DAP3 | 8494 | 0.455 | 0.2618 | No | ||
| 101 | GTF2A1 | 8677 | 0.379 | 0.2540 | No | ||
| 102 | ZPBP2 | 8684 | 0.376 | 0.2542 | No | ||
| 103 | ODZ1 | 9020 | 0.224 | 0.2392 | No | ||
| 104 | WNT3 | 9060 | 0.205 | 0.2377 | No | ||
| 105 | GFRA3 | 9172 | 0.162 | 0.2328 | No | ||
| 106 | IL17B | 9310 | 0.101 | 0.2266 | No | ||
| 107 | CCR3 | 9683 | -0.079 | 0.2097 | No | ||
| 108 | WDR34 | 10304 | -0.337 | 0.1816 | No | ||
| 109 | SEC23B | 10315 | -0.345 | 0.1817 | No | ||
| 110 | EPS8L1 | 10363 | -0.359 | 0.1800 | No | ||
| 111 | CDH20 | 10492 | -0.402 | 0.1747 | No | ||
| 112 | RAB8B | 11141 | -0.634 | 0.1458 | No | ||
| 113 | PPP2R2A | 11150 | -0.638 | 0.1463 | No | ||
| 114 | RALGDS | 11333 | -0.691 | 0.1389 | No | ||
| 115 | RPIA | 11453 | -0.733 | 0.1344 | No | ||
| 116 | GABRA1 | 11521 | -0.757 | 0.1324 | No | ||
| 117 | SGTB | 11640 | -0.796 | 0.1281 | No | ||
| 118 | PRIMA1 | 12216 | -0.981 | 0.1030 | No | ||
| 119 | HNRPC | 12282 | -1.002 | 0.1015 | No | ||
| 120 | SCN3A | 12680 | -1.117 | 0.0848 | No | ||
| 121 | SOX14 | 12979 | -1.209 | 0.0727 | No | ||
| 122 | CACNA2D3 | 13116 | -1.250 | 0.0682 | No | ||
| 123 | EN1 | 13659 | -1.411 | 0.0453 | No | ||
| 124 | CACNB2 | 13894 | -1.477 | 0.0366 | No | ||
| 125 | SPRY4 | 13917 | -1.483 | 0.0376 | No | ||
| 126 | ZZZ3 | 14196 | -1.566 | 0.0270 | No | ||
| 127 | DDX3X | 14411 | -1.623 | 0.0194 | No | ||
| 128 | PDE4D | 14460 | -1.636 | 0.0195 | No | ||
| 129 | KIAA1128 | 14665 | -1.701 | 0.0125 | No | ||
| 130 | FLRT3 | 14786 | -1.737 | 0.0094 | No | ||
| 131 | ID1 | 14941 | -1.781 | 0.0048 | No | ||
| 132 | NCAM2 | 15234 | -1.875 | -0.0060 | No | ||
| 133 | PPP2R2B | 15396 | -1.924 | -0.0108 | No | ||
| 134 | PHOX2B | 15429 | -1.933 | -0.0095 | No | ||
| 135 | LHX9 | 15468 | -1.946 | -0.0086 | No | ||
| 136 | SNAP25 | 15566 | -1.988 | -0.0103 | No | ||
| 137 | RHOJ | 15704 | -2.033 | -0.0138 | No | ||
| 138 | DDAH1 | 15823 | -2.070 | -0.0163 | No | ||
| 139 | PRKAG1 | 16085 | -2.147 | -0.0253 | No | ||
| 140 | MYO1E | 16592 | -2.321 | -0.0454 | No | ||
| 141 | CDH10 | 16631 | -2.333 | -0.0439 | No | ||
| 142 | MYL1 | 16742 | -2.372 | -0.0456 | No | ||
| 143 | CALD1 | 17060 | -2.487 | -0.0568 | No | ||
| 144 | GRIK2 | 17480 | -2.639 | -0.0723 | No | ||
| 145 | VSNL1 | 17748 | -2.748 | -0.0808 | No | ||
| 146 | ATP5G2 | 17774 | -2.762 | -0.0781 | No | ||
| 147 | SFRS3 | 17850 | -2.790 | -0.0777 | No | ||
| 148 | ZADH2 | 17870 | -2.796 | -0.0747 | No | ||
| 149 | PWWP2 | 17915 | -2.818 | -0.0728 | No | ||
| 150 | FOXP2 | 17925 | -2.821 | -0.0693 | No | ||
| 151 | ACTN1 | 18148 | -2.914 | -0.0755 | No | ||
| 152 | TCF4 | 18239 | -2.951 | -0.0755 | No | ||
| 153 | SLC25A13 | 18476 | -3.061 | -0.0821 | No | ||
| 154 | AP1G1 | 18539 | -3.091 | -0.0807 | No | ||
| 155 | FAF1 | 18567 | -3.102 | -0.0776 | No | ||
| 156 | ZFHX1B | 18599 | -3.124 | -0.0747 | No | ||
| 157 | TAL2 | 18615 | -3.135 | -0.0711 | No | ||
| 158 | MYF5 | 18846 | -3.261 | -0.0771 | No | ||
| 159 | SYNCRIP | 18869 | -3.273 | -0.0736 | No | ||
| 160 | CHCHD7 | 19061 | -3.388 | -0.0777 | No | ||
| 161 | ADK | 19431 | -3.635 | -0.0896 | No | ||
| 162 | DLX1 | 19783 | -3.932 | -0.1002 | No | ||
| 163 | INPPL1 | 19844 | -3.988 | -0.0975 | No | ||
| 164 | CXADR | 19874 | -4.015 | -0.0932 | No | ||
| 165 | STAG1 | 19878 | -4.015 | -0.0878 | No | ||
| 166 | DSTN | 20067 | -4.206 | -0.0906 | No | ||
| 167 | LMO6 | 20177 | -4.328 | -0.0896 | No | ||
| 168 | PEX7 | 20855 | -5.270 | -0.1134 | No | ||
| 169 | DUSP10 | 20883 | -5.300 | -0.1073 | No | ||
| 170 | CD68 | 21353 | -6.435 | -0.1199 | No | ||
| 171 | MATR3 | 21484 | -6.867 | -0.1164 | No | ||
| 172 | E2F3 | 21509 | -6.976 | -0.1078 | No | ||
| 173 | SLC25A4 | 21527 | -7.037 | -0.0989 | No | ||
| 174 | SFRS2 | 21702 | -8.098 | -0.0956 | No | ||
| 175 | TPM3 | 21741 | -8.392 | -0.0857 | No | ||
| 176 | TPM1 | 21827 | -9.610 | -0.0763 | No | ||
| 177 | NLN | 21828 | -9.662 | -0.0629 | No | ||
| 178 | CYR61 | 21874 | -11.031 | -0.0497 | No | ||
| 179 | PCBP4 | 21889 | -11.615 | -0.0342 | No | ||
| 180 | PBX3 | 21946 | -26.594 | 0.0000 | No |
