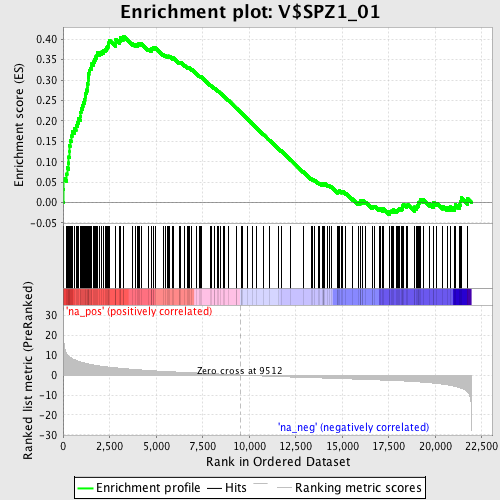
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$SPZ1_01 |
| Enrichment Score (ES) | 0.40765837 |
| Normalized Enrichment Score (NES) | 1.8318243 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009004305 |
| FWER p-Value | 0.095 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FGF13 | 12 | 20.624 | 0.0327 | Yes | ||
| 2 | SSBP2 | 36 | 16.873 | 0.0588 | Yes | ||
| 3 | HIF3A | 177 | 10.919 | 0.0700 | Yes | ||
| 4 | WDTC1 | 210 | 10.452 | 0.0854 | Yes | ||
| 5 | BAZ2A | 293 | 9.571 | 0.0970 | Yes | ||
| 6 | FOXJ2 | 307 | 9.460 | 0.1117 | Yes | ||
| 7 | TBCC | 339 | 9.185 | 0.1251 | Yes | ||
| 8 | PHF12 | 345 | 9.131 | 0.1395 | Yes | ||
| 9 | SRRM1 | 375 | 8.917 | 0.1526 | Yes | ||
| 10 | MYO1C | 442 | 8.465 | 0.1632 | Yes | ||
| 11 | TOM1L2 | 481 | 8.273 | 0.1748 | Yes | ||
| 12 | PPP1R10 | 600 | 7.677 | 0.1817 | Yes | ||
| 13 | NAB2 | 695 | 7.297 | 0.1892 | Yes | ||
| 14 | CRMP1 | 768 | 6.999 | 0.1972 | Yes | ||
| 15 | RBM12 | 827 | 6.796 | 0.2054 | Yes | ||
| 16 | CCND2 | 921 | 6.537 | 0.2117 | Yes | ||
| 17 | FBS1 | 937 | 6.504 | 0.2215 | Yes | ||
| 18 | DCTN1 | 965 | 6.422 | 0.2306 | Yes | ||
| 19 | STK35 | 1039 | 6.243 | 0.2373 | Yes | ||
| 20 | CNOT4 | 1072 | 6.146 | 0.2458 | Yes | ||
| 21 | PRKCBP1 | 1135 | 6.012 | 0.2526 | Yes | ||
| 22 | PPP2R5C | 1176 | 5.914 | 0.2603 | Yes | ||
| 23 | HIVEP1 | 1218 | 5.826 | 0.2678 | Yes | ||
| 24 | HIRA | 1246 | 5.757 | 0.2758 | Yes | ||
| 25 | AKT2 | 1306 | 5.599 | 0.2821 | Yes | ||
| 26 | ARNTL | 1310 | 5.592 | 0.2910 | Yes | ||
| 27 | MBD6 | 1339 | 5.529 | 0.2986 | Yes | ||
| 28 | CAMTA2 | 1345 | 5.516 | 0.3073 | Yes | ||
| 29 | MAZ | 1363 | 5.461 | 0.3153 | Yes | ||
| 30 | WHSC1L1 | 1392 | 5.411 | 0.3228 | Yes | ||
| 31 | NFKB2 | 1444 | 5.315 | 0.3290 | Yes | ||
| 32 | DDX17 | 1513 | 5.188 | 0.3342 | Yes | ||
| 33 | GNAI2 | 1546 | 5.136 | 0.3410 | Yes | ||
| 34 | SLC25A23 | 1647 | 4.978 | 0.3445 | Yes | ||
| 35 | MYLK | 1683 | 4.921 | 0.3508 | Yes | ||
| 36 | ILF3 | 1726 | 4.861 | 0.3567 | Yes | ||
| 37 | CHD4 | 1814 | 4.734 | 0.3603 | Yes | ||
| 38 | OSBPL7 | 1821 | 4.718 | 0.3676 | Yes | ||
| 39 | UBE2B | 1974 | 4.499 | 0.3679 | Yes | ||
| 40 | EIF4E | 2088 | 4.365 | 0.3698 | Yes | ||
| 41 | CPNE1 | 2187 | 4.234 | 0.3721 | Yes | ||
| 42 | TBC1D13 | 2259 | 4.160 | 0.3755 | Yes | ||
| 43 | ZFP91 | 2331 | 4.083 | 0.3788 | Yes | ||
| 44 | ABHD8 | 2392 | 4.018 | 0.3826 | Yes | ||
| 45 | NLK | 2430 | 3.973 | 0.3873 | Yes | ||
| 46 | ZBTB3 | 2460 | 3.941 | 0.3923 | Yes | ||
| 47 | PICALM | 2482 | 3.908 | 0.3976 | Yes | ||
| 48 | STK4 | 2809 | 3.603 | 0.3884 | Yes | ||
| 49 | CDK6 | 2833 | 3.585 | 0.3932 | Yes | ||
| 50 | TIAM1 | 2834 | 3.585 | 0.3989 | Yes | ||
| 51 | PCF11 | 3029 | 3.410 | 0.3955 | Yes | ||
| 52 | RBMS3 | 3065 | 3.379 | 0.3994 | Yes | ||
| 53 | BCL11A | 3071 | 3.369 | 0.4046 | Yes | ||
| 54 | SEC24B | 3221 | 3.243 | 0.4029 | Yes | ||
| 55 | LDB1 | 3233 | 3.237 | 0.4077 | Yes | ||
| 56 | UBAP1 | 3739 | 2.848 | 0.3890 | No | ||
| 57 | MAP1A | 3878 | 2.742 | 0.3871 | No | ||
| 58 | IGSF4D | 3994 | 2.651 | 0.3861 | No | ||
| 59 | CTCF | 4032 | 2.627 | 0.3886 | No | ||
| 60 | RNF12 | 4087 | 2.597 | 0.3904 | No | ||
| 61 | PITPNC1 | 4201 | 2.506 | 0.3892 | No | ||
| 62 | RASGEF1A | 4590 | 2.268 | 0.3750 | No | ||
| 63 | AMIGO2 | 4722 | 2.189 | 0.3725 | No | ||
| 64 | KCNK2 | 4761 | 2.164 | 0.3743 | No | ||
| 65 | EPN3 | 4767 | 2.161 | 0.3775 | No | ||
| 66 | MLLT7 | 4844 | 2.127 | 0.3775 | No | ||
| 67 | VCAM1 | 4861 | 2.117 | 0.3801 | No | ||
| 68 | DPYSL2 | 4962 | 2.060 | 0.3789 | No | ||
| 69 | SP1 | 5374 | 1.845 | 0.3630 | No | ||
| 70 | KCNK4 | 5495 | 1.781 | 0.3603 | No | ||
| 71 | SP4 | 5592 | 1.731 | 0.3587 | No | ||
| 72 | HDAC8 | 5645 | 1.704 | 0.3591 | No | ||
| 73 | CYLD | 5710 | 1.680 | 0.3588 | No | ||
| 74 | EPS15 | 5903 | 1.589 | 0.3526 | No | ||
| 75 | ADRB1 | 5918 | 1.585 | 0.3545 | No | ||
| 76 | BLOC1S3 | 6235 | 1.435 | 0.3423 | No | ||
| 77 | HMGN2 | 6245 | 1.428 | 0.3442 | No | ||
| 78 | PAX8 | 6298 | 1.406 | 0.3440 | No | ||
| 79 | NCDN | 6518 | 1.301 | 0.3361 | No | ||
| 80 | HOXC10 | 6671 | 1.224 | 0.3311 | No | ||
| 81 | SIGIRR | 6753 | 1.192 | 0.3293 | No | ||
| 82 | FOS | 6777 | 1.183 | 0.3301 | No | ||
| 83 | F13A1 | 6912 | 1.117 | 0.3258 | No | ||
| 84 | KCNC1 | 7142 | 1.020 | 0.3169 | No | ||
| 85 | FHL3 | 7333 | 0.936 | 0.3097 | No | ||
| 86 | KCNV2 | 7387 | 0.915 | 0.3087 | No | ||
| 87 | FXYD4 | 7423 | 0.899 | 0.3085 | No | ||
| 88 | ADNP | 7904 | 0.701 | 0.2876 | No | ||
| 89 | POU2F1 | 7999 | 0.663 | 0.2844 | No | ||
| 90 | SP7 | 8112 | 0.614 | 0.2802 | No | ||
| 91 | ZRANB1 | 8131 | 0.604 | 0.2804 | No | ||
| 92 | ECEL1 | 8268 | 0.551 | 0.2750 | No | ||
| 93 | HYAL2 | 8338 | 0.521 | 0.2727 | No | ||
| 94 | SOX10 | 8462 | 0.470 | 0.2678 | No | ||
| 95 | CDK5R2 | 8629 | 0.396 | 0.2608 | No | ||
| 96 | GTF2A1 | 8677 | 0.379 | 0.2592 | No | ||
| 97 | SLC17A7 | 8882 | 0.290 | 0.2503 | No | ||
| 98 | IL17B | 9310 | 0.101 | 0.2309 | No | ||
| 99 | STAC2 | 9317 | 0.099 | 0.2308 | No | ||
| 100 | HOXB8 | 9566 | -0.026 | 0.2194 | No | ||
| 101 | IRX5 | 9571 | -0.029 | 0.2193 | No | ||
| 102 | WNT10A | 9640 | -0.061 | 0.2162 | No | ||
| 103 | HOXB6 | 9880 | -0.167 | 0.2055 | No | ||
| 104 | PPP1R1B | 10155 | -0.280 | 0.1934 | No | ||
| 105 | STAG2 | 10386 | -0.368 | 0.1834 | No | ||
| 106 | CASKIN2 | 10778 | -0.506 | 0.1663 | No | ||
| 107 | TENC1 | 11086 | -0.611 | 0.1531 | No | ||
| 108 | HOXB9 | 11583 | -0.779 | 0.1316 | No | ||
| 109 | NPPA | 11711 | -0.829 | 0.1271 | No | ||
| 110 | TPT1 | 12224 | -0.983 | 0.1052 | No | ||
| 111 | MOV10 | 12911 | -1.190 | 0.0756 | No | ||
| 112 | SHH | 13338 | -1.311 | 0.0581 | No | ||
| 113 | POU3F3 | 13418 | -1.338 | 0.0566 | No | ||
| 114 | RIMS1 | 13506 | -1.365 | 0.0548 | No | ||
| 115 | ACCN2 | 13707 | -1.429 | 0.0480 | No | ||
| 116 | PGR | 13802 | -1.451 | 0.0460 | No | ||
| 117 | SPRY4 | 13917 | -1.483 | 0.0431 | No | ||
| 118 | SOX15 | 13972 | -1.497 | 0.0431 | No | ||
| 119 | HSD3B7 | 13979 | -1.500 | 0.0452 | No | ||
| 120 | EPHB1 | 13989 | -1.503 | 0.0472 | No | ||
| 121 | EIF4G1 | 14061 | -1.526 | 0.0464 | No | ||
| 122 | PSD | 14229 | -1.574 | 0.0413 | No | ||
| 123 | HOXB1 | 14294 | -1.594 | 0.0409 | No | ||
| 124 | S100A4 | 14405 | -1.622 | 0.0385 | No | ||
| 125 | HR | 14740 | -1.724 | 0.0259 | No | ||
| 126 | UQCRH | 14758 | -1.729 | 0.0279 | No | ||
| 127 | CALM2 | 14820 | -1.748 | 0.0279 | No | ||
| 128 | TCF2 | 14838 | -1.752 | 0.0300 | No | ||
| 129 | HOXB4 | 14983 | -1.795 | 0.0262 | No | ||
| 130 | FGF17 | 15033 | -1.809 | 0.0269 | No | ||
| 131 | HOXA9 | 15182 | -1.861 | 0.0231 | No | ||
| 132 | GLRA1 | 15559 | -1.986 | 0.0090 | No | ||
| 133 | KCNN2 | 15845 | -2.077 | -0.0007 | No | ||
| 134 | MAGED2 | 15954 | -2.109 | -0.0023 | No | ||
| 135 | XPO7 | 15957 | -2.111 | 0.0010 | No | ||
| 136 | SOX3 | 15963 | -2.112 | 0.0042 | No | ||
| 137 | PRKAG1 | 16085 | -2.147 | 0.0021 | No | ||
| 138 | SUMO2 | 16099 | -2.150 | 0.0050 | No | ||
| 139 | ADAMTSL1 | 16264 | -2.207 | 0.0010 | No | ||
| 140 | SLITRK1 | 16616 | -2.328 | -0.0114 | No | ||
| 141 | HOXA10 | 16650 | -2.337 | -0.0091 | No | ||
| 142 | SHANK1 | 16727 | -2.368 | -0.0088 | No | ||
| 143 | EIF4G2 | 16984 | -2.461 | -0.0166 | No | ||
| 144 | BDNF | 17040 | -2.479 | -0.0151 | No | ||
| 145 | NTRK3 | 17176 | -2.523 | -0.0173 | No | ||
| 146 | CUL2 | 17215 | -2.540 | -0.0149 | No | ||
| 147 | GRK5 | 17526 | -2.656 | -0.0249 | No | ||
| 148 | HOXD9 | 17534 | -2.658 | -0.0209 | No | ||
| 149 | GREM1 | 17620 | -2.689 | -0.0205 | No | ||
| 150 | FST | 17720 | -2.736 | -0.0206 | No | ||
| 151 | ADAMTS9 | 17754 | -2.753 | -0.0177 | No | ||
| 152 | GPM6B | 17903 | -2.815 | -0.0200 | No | ||
| 153 | PCSK2 | 17967 | -2.840 | -0.0183 | No | ||
| 154 | APBA2BP | 18016 | -2.862 | -0.0159 | No | ||
| 155 | BCL7A | 18062 | -2.882 | -0.0133 | No | ||
| 156 | DQX1 | 18177 | -2.928 | -0.0138 | No | ||
| 157 | HMGA1 | 18236 | -2.950 | -0.0117 | No | ||
| 158 | PRRX1 | 18256 | -2.957 | -0.0078 | No | ||
| 159 | SMARCC1 | 18284 | -2.969 | -0.0043 | No | ||
| 160 | MRC2 | 18450 | -3.051 | -0.0070 | No | ||
| 161 | ATF3 | 18527 | -3.087 | -0.0055 | No | ||
| 162 | SYNCRIP | 18869 | -3.273 | -0.0159 | No | ||
| 163 | HTF9C | 18872 | -3.275 | -0.0107 | No | ||
| 164 | CNTFR | 19002 | -3.356 | -0.0112 | No | ||
| 165 | KPNB1 | 19033 | -3.372 | -0.0072 | No | ||
| 166 | EFEMP2 | 19088 | -3.407 | -0.0041 | No | ||
| 167 | SIX4 | 19099 | -3.414 | 0.0009 | No | ||
| 168 | SCRT2 | 19174 | -3.467 | 0.0031 | No | ||
| 169 | XPO1 | 19211 | -3.485 | 0.0071 | No | ||
| 170 | YTHDF2 | 19339 | -3.567 | 0.0070 | No | ||
| 171 | MRPL40 | 19681 | -3.832 | -0.0025 | No | ||
| 172 | GATA3 | 19901 | -4.038 | -0.0061 | No | ||
| 173 | DLGAP4 | 19907 | -4.040 | 0.0002 | No | ||
| 174 | PSCD3 | 20086 | -4.223 | -0.0012 | No | ||
| 175 | PABPC1 | 20409 | -4.595 | -0.0086 | No | ||
| 176 | NR2F2 | 20641 | -4.901 | -0.0113 | No | ||
| 177 | LMO4 | 20804 | -5.199 | -0.0103 | No | ||
| 178 | CXXC4 | 21019 | -5.527 | -0.0113 | No | ||
| 179 | RANBP1 | 21068 | -5.642 | -0.0044 | No | ||
| 180 | SNRPD1 | 21298 | -6.287 | -0.0048 | No | ||
| 181 | MRPS18B | 21359 | -6.463 | 0.0029 | No | ||
| 182 | CGGBP1 | 21380 | -6.513 | 0.0125 | No | ||
| 183 | TPM3 | 21741 | -8.392 | 0.0095 | No |
