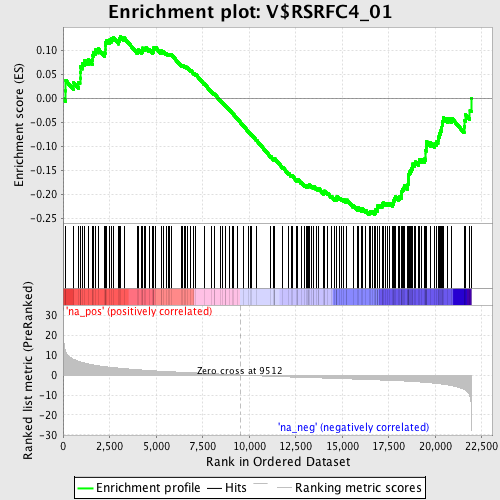
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$RSRFC4_01 |
| Enrichment Score (ES) | -0.24208064 |
| Normalized Enrichment Score (NES) | -1.1875021 |
| Nominal p-value | 0.10240964 |
| FDR q-value | 0.4490251 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLCO3A1 | 139 | 11.666 | 0.0158 | No | ||
| 2 | CENTD1 | 147 | 11.538 | 0.0373 | No | ||
| 3 | CAPN3 | 572 | 7.819 | 0.0327 | No | ||
| 4 | ATP2A3 | 840 | 6.761 | 0.0333 | No | ||
| 5 | CUTL1 | 908 | 6.567 | 0.0427 | No | ||
| 6 | EDG1 | 914 | 6.554 | 0.0549 | No | ||
| 7 | TOB2 | 930 | 6.514 | 0.0666 | No | ||
| 8 | FOXP1 | 1045 | 6.223 | 0.0731 | No | ||
| 9 | ACTA1 | 1163 | 5.929 | 0.0790 | No | ||
| 10 | MGST3 | 1353 | 5.500 | 0.0808 | No | ||
| 11 | FBXW11 | 1563 | 5.105 | 0.0809 | No | ||
| 12 | LUC7L | 1592 | 5.065 | 0.0892 | No | ||
| 13 | NFAT5 | 1636 | 4.993 | 0.0967 | No | ||
| 14 | ALS2CR2 | 1735 | 4.849 | 0.1014 | No | ||
| 15 | CDC42EP3 | 1880 | 4.634 | 0.1036 | No | ||
| 16 | NR4A1 | 2240 | 4.181 | 0.0950 | No | ||
| 17 | LZTS2 | 2258 | 4.161 | 0.1021 | No | ||
| 18 | CFL2 | 2263 | 4.155 | 0.1098 | No | ||
| 19 | TMEPAI | 2276 | 4.142 | 0.1171 | No | ||
| 20 | SLAMF1 | 2349 | 4.066 | 0.1216 | No | ||
| 21 | PLAGL2 | 2501 | 3.889 | 0.1220 | No | ||
| 22 | CASQ1 | 2600 | 3.808 | 0.1247 | No | ||
| 23 | PANK1 | 2708 | 3.699 | 0.1268 | No | ||
| 24 | WSB2 | 3001 | 3.433 | 0.1199 | No | ||
| 25 | TRPS1 | 3024 | 3.417 | 0.1254 | No | ||
| 26 | RBMS3 | 3065 | 3.379 | 0.1300 | No | ||
| 27 | SMARCA2 | 3274 | 3.202 | 0.1265 | No | ||
| 28 | NEDD4 | 3996 | 2.649 | 0.0984 | No | ||
| 29 | SSH3 | 4039 | 2.621 | 0.1014 | No | ||
| 30 | CPEB4 | 4224 | 2.491 | 0.0977 | No | ||
| 31 | ZHX2 | 4253 | 2.468 | 0.1011 | No | ||
| 32 | AMPD1 | 4265 | 2.463 | 0.1053 | No | ||
| 33 | HCFC1R1 | 4387 | 2.393 | 0.1043 | No | ||
| 34 | PTPN1 | 4451 | 2.352 | 0.1058 | No | ||
| 35 | ATP1B2 | 4623 | 2.244 | 0.1022 | No | ||
| 36 | ARHGAP26 | 4808 | 2.142 | 0.0979 | No | ||
| 37 | ITGB3BP | 4840 | 2.129 | 0.1005 | No | ||
| 38 | PAK6 | 4864 | 2.116 | 0.1034 | No | ||
| 39 | DTNB | 4877 | 2.109 | 0.1069 | No | ||
| 40 | MRPS23 | 4965 | 2.059 | 0.1068 | No | ||
| 41 | HRASLS | 5270 | 1.892 | 0.0964 | No | ||
| 42 | GRIN2B | 5286 | 1.882 | 0.0993 | No | ||
| 43 | KCNQ5 | 5382 | 1.841 | 0.0984 | No | ||
| 44 | EHD1 | 5548 | 1.750 | 0.0942 | No | ||
| 45 | HAO1 | 5666 | 1.696 | 0.0920 | No | ||
| 46 | SH3GLB2 | 5722 | 1.675 | 0.0927 | No | ||
| 47 | RICS | 5811 | 1.631 | 0.0917 | No | ||
| 48 | KBTBD10 | 6349 | 1.384 | 0.0697 | No | ||
| 49 | RG9MTD2 | 6405 | 1.359 | 0.0697 | No | ||
| 50 | DNAJA4 | 6526 | 1.297 | 0.0667 | No | ||
| 51 | WFDC1 | 6591 | 1.263 | 0.0661 | No | ||
| 52 | CASQ2 | 6661 | 1.228 | 0.0653 | No | ||
| 53 | SMCR7 | 6859 | 1.146 | 0.0584 | No | ||
| 54 | CYP2E1 | 7027 | 1.068 | 0.0528 | No | ||
| 55 | MAP2K5 | 7135 | 1.023 | 0.0498 | No | ||
| 56 | PRDM1 | 7584 | 0.833 | 0.0308 | No | ||
| 57 | PPP1R3D | 7966 | 0.675 | 0.0146 | No | ||
| 58 | USH3A | 8122 | 0.610 | 0.0086 | No | ||
| 59 | SLC26A6 | 8130 | 0.606 | 0.0094 | No | ||
| 60 | MEOX2 | 8437 | 0.480 | -0.0037 | No | ||
| 61 | SMPX | 8545 | 0.434 | -0.0078 | No | ||
| 62 | TNFRSF17 | 8734 | 0.347 | -0.0158 | No | ||
| 63 | TNNC1 | 8744 | 0.343 | -0.0156 | No | ||
| 64 | MYOZ2 | 8966 | 0.249 | -0.0252 | No | ||
| 65 | CLDN23 | 9088 | 0.195 | -0.0304 | No | ||
| 66 | MUSK | 9181 | 0.160 | -0.0344 | No | ||
| 67 | PPP2R3A | 9371 | 0.071 | -0.0429 | No | ||
| 68 | GPR153 | 9687 | -0.080 | -0.0572 | No | ||
| 69 | DLX5 | 9964 | -0.203 | -0.0695 | No | ||
| 70 | POU4F2 | 10093 | -0.260 | -0.0749 | No | ||
| 71 | RALY | 10117 | -0.266 | -0.0755 | No | ||
| 72 | STAG2 | 10386 | -0.368 | -0.0871 | No | ||
| 73 | ADCY2 | 11164 | -0.644 | -0.1216 | No | ||
| 74 | HOXC4 | 11324 | -0.689 | -0.1276 | No | ||
| 75 | PCDH11X | 11340 | -0.694 | -0.1269 | No | ||
| 76 | ANKMY2 | 11350 | -0.696 | -0.1260 | No | ||
| 77 | KBTBD5 | 11365 | -0.703 | -0.1253 | No | ||
| 78 | CAV3 | 11814 | -0.860 | -0.1443 | No | ||
| 79 | RAP2C | 12125 | -0.957 | -0.1567 | No | ||
| 80 | NDRG2 | 12273 | -1.000 | -0.1616 | No | ||
| 81 | KLF14 | 12304 | -1.008 | -0.1611 | No | ||
| 82 | LRRTM4 | 12556 | -1.084 | -0.1705 | No | ||
| 83 | ADAM11 | 12585 | -1.091 | -0.1697 | No | ||
| 84 | HOXD4 | 12616 | -1.101 | -0.1690 | No | ||
| 85 | MYL2 | 12787 | -1.155 | -0.1746 | No | ||
| 86 | SLC8A3 | 12992 | -1.215 | -0.1817 | No | ||
| 87 | CSRP3 | 13091 | -1.242 | -0.1839 | No | ||
| 88 | CACNA2D3 | 13116 | -1.250 | -0.1826 | No | ||
| 89 | IRS1 | 13166 | -1.265 | -0.1824 | No | ||
| 90 | SMYD1 | 13201 | -1.275 | -0.1816 | No | ||
| 91 | GPC4 | 13223 | -1.281 | -0.1801 | No | ||
| 92 | ZFPM2 | 13347 | -1.312 | -0.1833 | No | ||
| 93 | FILIP1 | 13437 | -1.344 | -0.1848 | No | ||
| 94 | DKK2 | 13476 | -1.354 | -0.1840 | No | ||
| 95 | LIF | 13625 | -1.399 | -0.1881 | No | ||
| 96 | HFE2 | 13710 | -1.430 | -0.1893 | No | ||
| 97 | RASGEF1B | 13738 | -1.437 | -0.1878 | No | ||
| 98 | TYRO3 | 14002 | -1.507 | -0.1970 | No | ||
| 99 | CYP26B1 | 14035 | -1.518 | -0.1956 | No | ||
| 100 | INPP4A | 14043 | -1.520 | -0.1930 | No | ||
| 101 | TCEA3 | 14208 | -1.570 | -0.1976 | No | ||
| 102 | S100A4 | 14405 | -1.622 | -0.2035 | No | ||
| 103 | GCK | 14607 | -1.683 | -0.2096 | No | ||
| 104 | SV2A | 14689 | -1.708 | -0.2100 | No | ||
| 105 | ASB16 | 14703 | -1.713 | -0.2074 | No | ||
| 106 | TNNC2 | 14714 | -1.715 | -0.2046 | No | ||
| 107 | RTBDN | 14869 | -1.761 | -0.2083 | No | ||
| 108 | HOXB4 | 14983 | -1.795 | -0.2101 | No | ||
| 109 | PTPRO | 15085 | -1.829 | -0.2113 | No | ||
| 110 | CLCN1 | 15203 | -1.866 | -0.2131 | No | ||
| 111 | PDLIM1 | 15253 | -1.882 | -0.2118 | No | ||
| 112 | RASGRP3 | 15577 | -1.992 | -0.2229 | No | ||
| 113 | TTYH2 | 15836 | -2.074 | -0.2308 | No | ||
| 114 | ARHGEF15 | 15857 | -2.081 | -0.2277 | No | ||
| 115 | MYL3 | 16041 | -2.134 | -0.2321 | No | ||
| 116 | PRKAG1 | 16085 | -2.147 | -0.2300 | No | ||
| 117 | ESR1 | 16252 | -2.203 | -0.2334 | No | ||
| 118 | HAPLN1 | 16441 | -2.267 | -0.2378 | Yes | ||
| 119 | RAB2 | 16500 | -2.289 | -0.2361 | Yes | ||
| 120 | HAS2 | 16601 | -2.323 | -0.2363 | Yes | ||
| 121 | SHANK1 | 16727 | -2.368 | -0.2375 | Yes | ||
| 122 | DMD | 16796 | -2.392 | -0.2361 | Yes | ||
| 123 | ADCYAP1 | 16799 | -2.394 | -0.2317 | Yes | ||
| 124 | FGF12 | 16906 | -2.433 | -0.2319 | Yes | ||
| 125 | ITGA7 | 16908 | -2.433 | -0.2273 | Yes | ||
| 126 | ADAM23 | 16913 | -2.436 | -0.2229 | Yes | ||
| 127 | NRXN3 | 17018 | -2.473 | -0.2230 | Yes | ||
| 128 | COL8A1 | 17137 | -2.511 | -0.2236 | Yes | ||
| 129 | EEF1A2 | 17157 | -2.518 | -0.2197 | Yes | ||
| 130 | MEF2C | 17220 | -2.542 | -0.2178 | Yes | ||
| 131 | CSPG3 | 17348 | -2.597 | -0.2187 | Yes | ||
| 132 | USP2 | 17454 | -2.632 | -0.2185 | Yes | ||
| 133 | NR2F1 | 17563 | -2.666 | -0.2184 | Yes | ||
| 134 | FST | 17720 | -2.736 | -0.2204 | Yes | ||
| 135 | LRRC20 | 17743 | -2.746 | -0.2162 | Yes | ||
| 136 | CUL3 | 17767 | -2.760 | -0.2120 | Yes | ||
| 137 | EPHA7 | 17815 | -2.779 | -0.2089 | Yes | ||
| 138 | TNNI3K | 17847 | -2.789 | -0.2050 | Yes | ||
| 139 | CLDN14 | 18035 | -2.870 | -0.2081 | Yes | ||
| 140 | CNTN1 | 18087 | -2.892 | -0.2050 | Yes | ||
| 141 | MYH3 | 18167 | -2.923 | -0.2031 | Yes | ||
| 142 | THBS2 | 18184 | -2.930 | -0.1982 | Yes | ||
| 143 | HDAC9 | 18201 | -2.936 | -0.1934 | Yes | ||
| 144 | ARR3 | 18230 | -2.948 | -0.1891 | Yes | ||
| 145 | PCDH9 | 18304 | -2.978 | -0.1868 | Yes | ||
| 146 | ITSN1 | 18327 | -2.991 | -0.1821 | Yes | ||
| 147 | POU4F1 | 18516 | -3.081 | -0.1849 | Yes | ||
| 148 | ATF3 | 18527 | -3.087 | -0.1795 | Yes | ||
| 149 | MBNL2 | 18541 | -3.091 | -0.1742 | Yes | ||
| 150 | TPP2 | 18549 | -3.093 | -0.1687 | Yes | ||
| 151 | TWIST1 | 18561 | -3.099 | -0.1633 | Yes | ||
| 152 | TRDN | 18571 | -3.102 | -0.1578 | Yes | ||
| 153 | SLC9A5 | 18601 | -3.127 | -0.1532 | Yes | ||
| 154 | RGS3 | 18672 | -3.160 | -0.1504 | Yes | ||
| 155 | PAX3 | 18724 | -3.198 | -0.1467 | Yes | ||
| 156 | CKM | 18765 | -3.221 | -0.1424 | Yes | ||
| 157 | BAI3 | 18774 | -3.223 | -0.1367 | Yes | ||
| 158 | PYY | 18885 | -3.286 | -0.1355 | Yes | ||
| 159 | STAC | 18953 | -3.327 | -0.1323 | Yes | ||
| 160 | ELMO1 | 19121 | -3.431 | -0.1334 | Yes | ||
| 161 | RASGRF1 | 19135 | -3.440 | -0.1275 | Yes | ||
| 162 | LECT1 | 19259 | -3.513 | -0.1265 | Yes | ||
| 163 | STOML2 | 19403 | -3.613 | -0.1262 | Yes | ||
| 164 | SOX5 | 19470 | -3.663 | -0.1223 | Yes | ||
| 165 | DGKI | 19471 | -3.663 | -0.1153 | Yes | ||
| 166 | TNNI2 | 19472 | -3.664 | -0.1084 | Yes | ||
| 167 | DIRAS1 | 19507 | -3.694 | -0.1029 | Yes | ||
| 168 | WBP4 | 19517 | -3.702 | -0.0963 | Yes | ||
| 169 | PIK3R3 | 19528 | -3.708 | -0.0897 | Yes | ||
| 170 | KCNN1 | 19743 | -3.892 | -0.0922 | Yes | ||
| 171 | ELAVL4 | 19973 | -4.101 | -0.0949 | Yes | ||
| 172 | CTNND2 | 20052 | -4.197 | -0.0905 | Yes | ||
| 173 | EYA1 | 20145 | -4.286 | -0.0866 | Yes | ||
| 174 | BNIP3 | 20187 | -4.336 | -0.0803 | Yes | ||
| 175 | ART5 | 20215 | -4.372 | -0.0732 | Yes | ||
| 176 | PDGFRA | 20279 | -4.427 | -0.0677 | Yes | ||
| 177 | EXTL1 | 20324 | -4.478 | -0.0612 | Yes | ||
| 178 | BRUNOL4 | 20358 | -4.522 | -0.0541 | Yes | ||
| 179 | LCP1 | 20395 | -4.572 | -0.0471 | Yes | ||
| 180 | ENO3 | 20436 | -4.643 | -0.0401 | Yes | ||
| 181 | KPNA3 | 20663 | -4.935 | -0.0411 | Yes | ||
| 182 | MAB21L2 | 20881 | -5.298 | -0.0411 | Yes | ||
| 183 | CRTAP | 21546 | -7.110 | -0.0581 | Yes | ||
| 184 | KTN1 | 21583 | -7.244 | -0.0460 | Yes | ||
| 185 | SIPA1L1 | 21621 | -7.482 | -0.0335 | Yes | ||
| 186 | BZW2 | 21862 | -10.683 | -0.0242 | Yes | ||
| 187 | TIMP2 | 21931 | -14.789 | 0.0007 | Yes |
