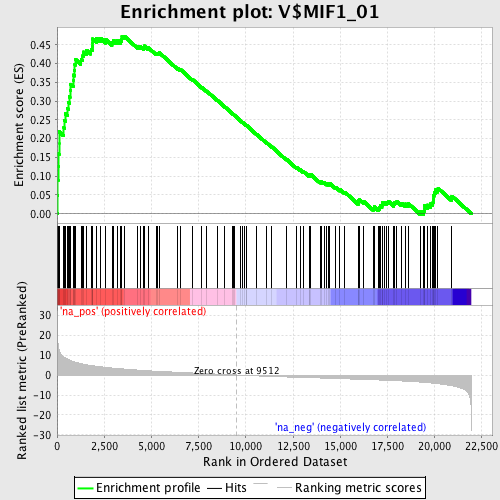
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MIF1_01 |
| Enrichment Score (ES) | 0.47346154 |
| Normalized Enrichment Score (NES) | 2.0259259 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0054941718 |
| FWER p-Value | 0.012 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FGF13 | 12 | 20.624 | 0.0487 | Yes | ||
| 2 | SOCS1 | 32 | 17.187 | 0.0889 | Yes | ||
| 3 | BCL2L1 | 52 | 15.577 | 0.1252 | Yes | ||
| 4 | DHRS3 | 61 | 14.736 | 0.1601 | Yes | ||
| 5 | CCRK | 106 | 12.699 | 0.1884 | Yes | ||
| 6 | CBX4 | 109 | 12.686 | 0.2186 | Yes | ||
| 7 | RNF25 | 358 | 9.027 | 0.2288 | Yes | ||
| 8 | YWHAE | 396 | 8.835 | 0.2482 | Yes | ||
| 9 | MYO1C | 442 | 8.465 | 0.2664 | Yes | ||
| 10 | TMEM24 | 550 | 7.916 | 0.2804 | Yes | ||
| 11 | DHX30 | 590 | 7.724 | 0.2971 | Yes | ||
| 12 | RNF41 | 647 | 7.518 | 0.3125 | Yes | ||
| 13 | DDX31 | 685 | 7.344 | 0.3283 | Yes | ||
| 14 | MADD | 698 | 7.257 | 0.3451 | Yes | ||
| 15 | SPRY2 | 850 | 6.721 | 0.3542 | Yes | ||
| 16 | RFX2 | 859 | 6.698 | 0.3699 | Yes | ||
| 17 | CCND2 | 921 | 6.537 | 0.3827 | Yes | ||
| 18 | CCDC6 | 941 | 6.492 | 0.3973 | Yes | ||
| 19 | DCTN1 | 965 | 6.422 | 0.4116 | Yes | ||
| 20 | NCOA5 | 1265 | 5.709 | 0.4116 | Yes | ||
| 21 | SREBF1 | 1355 | 5.494 | 0.4206 | Yes | ||
| 22 | AP1M1 | 1406 | 5.387 | 0.4312 | Yes | ||
| 23 | NEDD8 | 1564 | 5.103 | 0.4362 | Yes | ||
| 24 | PTDSS2 | 1795 | 4.760 | 0.4370 | Yes | ||
| 25 | PSCD2 | 1860 | 4.668 | 0.4452 | Yes | ||
| 26 | CBLB | 1885 | 4.615 | 0.4552 | Yes | ||
| 27 | CBX6 | 1891 | 4.608 | 0.4659 | Yes | ||
| 28 | E2F5 | 2082 | 4.371 | 0.4677 | Yes | ||
| 29 | HM13 | 2285 | 4.127 | 0.4683 | Yes | ||
| 30 | DNAJC1 | 2580 | 3.829 | 0.4640 | Yes | ||
| 31 | TBPL1 | 2909 | 3.510 | 0.4573 | Yes | ||
| 32 | TRIM39 | 2994 | 3.442 | 0.4617 | Yes | ||
| 33 | ARL4A | 3173 | 3.289 | 0.4614 | Yes | ||
| 34 | TPCN1 | 3344 | 3.140 | 0.4611 | Yes | ||
| 35 | NEDD9 | 3408 | 3.097 | 0.4656 | Yes | ||
| 36 | NEK2 | 3436 | 3.076 | 0.4718 | Yes | ||
| 37 | KIF3B | 3555 | 2.976 | 0.4735 | Yes | ||
| 38 | POLD4 | 4275 | 2.455 | 0.4464 | No | ||
| 39 | SNF1LK | 4417 | 2.374 | 0.4456 | No | ||
| 40 | RASGEF1A | 4590 | 2.268 | 0.4432 | No | ||
| 41 | VEGF | 4617 | 2.251 | 0.4473 | No | ||
| 42 | GTF3C4 | 4832 | 2.134 | 0.4426 | No | ||
| 43 | DPAGT1 | 5255 | 1.897 | 0.4278 | No | ||
| 44 | ADCK4 | 5341 | 1.857 | 0.4284 | No | ||
| 45 | GMPR2 | 5417 | 1.823 | 0.4293 | No | ||
| 46 | WARS2 | 6391 | 1.368 | 0.3880 | No | ||
| 47 | NCDN | 6518 | 1.301 | 0.3853 | No | ||
| 48 | LRP2 | 7184 | 1.004 | 0.3573 | No | ||
| 49 | SALL2 | 7672 | 0.796 | 0.3369 | No | ||
| 50 | CSMD3 | 7920 | 0.696 | 0.3272 | No | ||
| 51 | SPA17 | 8474 | 0.464 | 0.3030 | No | ||
| 52 | MAGED1 | 8886 | 0.288 | 0.2849 | No | ||
| 53 | ITGB8 | 9268 | 0.123 | 0.2677 | No | ||
| 54 | GPRC5D | 9323 | 0.097 | 0.2655 | No | ||
| 55 | NRGN | 9388 | 0.064 | 0.2627 | No | ||
| 56 | EFNB3 | 9743 | -0.106 | 0.2468 | No | ||
| 57 | MRPL10 | 9818 | -0.141 | 0.2437 | No | ||
| 58 | STMN4 | 9951 | -0.199 | 0.2381 | No | ||
| 59 | ARID1A | 10021 | -0.231 | 0.2355 | No | ||
| 60 | PRKCSH | 10057 | -0.246 | 0.2345 | No | ||
| 61 | LSM7 | 10560 | -0.425 | 0.2125 | No | ||
| 62 | TGFB3 | 11105 | -0.622 | 0.1891 | No | ||
| 63 | TTC8 | 11372 | -0.706 | 0.1786 | No | ||
| 64 | BBS2 | 12147 | -0.960 | 0.1455 | No | ||
| 65 | GLRA3 | 12704 | -1.124 | 0.1227 | No | ||
| 66 | CPEB2 | 12870 | -1.178 | 0.1179 | No | ||
| 67 | TTC16 | 13072 | -1.237 | 0.1117 | No | ||
| 68 | BMP6 | 13373 | -1.323 | 0.1011 | No | ||
| 69 | PLA2G12A | 13377 | -1.325 | 0.1041 | No | ||
| 70 | FILIP1 | 13437 | -1.344 | 0.1046 | No | ||
| 71 | PACRG | 13938 | -1.487 | 0.0853 | No | ||
| 72 | DNAH9 | 14023 | -1.516 | 0.0851 | No | ||
| 73 | SDCBP2 | 14147 | -1.549 | 0.0831 | No | ||
| 74 | KIF17 | 14293 | -1.594 | 0.0803 | No | ||
| 75 | PLCB3 | 14401 | -1.621 | 0.0793 | No | ||
| 76 | PARK2 | 14444 | -1.631 | 0.0813 | No | ||
| 77 | HMGCS1 | 14755 | -1.729 | 0.0712 | No | ||
| 78 | TUSC3 | 14989 | -1.798 | 0.0648 | No | ||
| 79 | GRIK5 | 15254 | -1.882 | 0.0572 | No | ||
| 80 | STK36 | 15953 | -2.109 | 0.0303 | No | ||
| 81 | SOX3 | 15963 | -2.112 | 0.0349 | No | ||
| 82 | GRM3 | 16010 | -2.125 | 0.0379 | No | ||
| 83 | TEKT2 | 16224 | -2.193 | 0.0334 | No | ||
| 84 | CARS | 16746 | -2.373 | 0.0152 | No | ||
| 85 | DMD | 16796 | -2.392 | 0.0187 | No | ||
| 86 | BDNF | 17040 | -2.479 | 0.0135 | No | ||
| 87 | GALNT14 | 17091 | -2.496 | 0.0171 | No | ||
| 88 | CD44 | 17112 | -2.503 | 0.0222 | No | ||
| 89 | EMX2 | 17217 | -2.542 | 0.0235 | No | ||
| 90 | CX3CL1 | 17229 | -2.546 | 0.0291 | No | ||
| 91 | ELAVL2 | 17342 | -2.594 | 0.0301 | No | ||
| 92 | MDH1B | 17457 | -2.633 | 0.0312 | No | ||
| 93 | FIBCD1 | 17544 | -2.660 | 0.0336 | No | ||
| 94 | SMAD5 | 17846 | -2.789 | 0.0265 | No | ||
| 95 | HAT1 | 17894 | -2.808 | 0.0311 | No | ||
| 96 | PHF15 | 18005 | -2.856 | 0.0328 | No | ||
| 97 | GRID2 | 18262 | -2.959 | 0.0282 | No | ||
| 98 | TLX3 | 18469 | -3.058 | 0.0261 | No | ||
| 99 | THNSL1 | 18602 | -3.127 | 0.0275 | No | ||
| 100 | PTCH2 | 19247 | -3.508 | 0.0064 | No | ||
| 101 | ADK | 19431 | -3.635 | 0.0067 | No | ||
| 102 | CRLF1 | 19462 | -3.657 | 0.0140 | No | ||
| 103 | SOX5 | 19470 | -3.663 | 0.0225 | No | ||
| 104 | GRWD1 | 19651 | -3.808 | 0.0233 | No | ||
| 105 | NAT5 | 19792 | -3.941 | 0.0263 | No | ||
| 106 | TBC1D16 | 19916 | -4.048 | 0.0304 | No | ||
| 107 | WWP1 | 19929 | -4.061 | 0.0395 | No | ||
| 108 | MDH1 | 19951 | -4.074 | 0.0483 | No | ||
| 109 | MYCBP | 19976 | -4.104 | 0.0570 | No | ||
| 110 | FGR | 20050 | -4.194 | 0.0637 | No | ||
| 111 | NELF | 20182 | -4.332 | 0.0680 | No | ||
| 112 | CHCHD4 | 20917 | -5.349 | 0.0472 | No |
