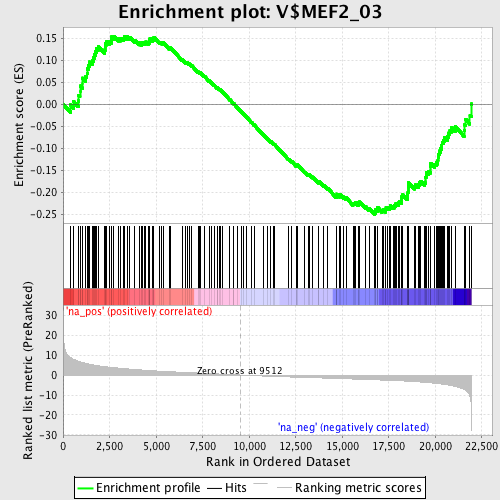
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MEF2_03 |
| Enrichment Score (ES) | -0.24983516 |
| Normalized Enrichment Score (NES) | -1.2221773 |
| Nominal p-value | 0.061797753 |
| FDR q-value | 0.38601544 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | POFUT1 | 399 | 8.796 | -0.0014 | No | ||
| 2 | CAPN3 | 572 | 7.819 | 0.0057 | No | ||
| 3 | SEMA7A | 817 | 6.836 | 0.0076 | No | ||
| 4 | ATP2A3 | 840 | 6.761 | 0.0196 | No | ||
| 5 | CUTL1 | 908 | 6.567 | 0.0291 | No | ||
| 6 | EDG1 | 914 | 6.554 | 0.0415 | No | ||
| 7 | RUNDC1 | 1043 | 6.228 | 0.0476 | No | ||
| 8 | FOXP1 | 1045 | 6.223 | 0.0595 | No | ||
| 9 | HIVEP1 | 1218 | 5.826 | 0.0627 | No | ||
| 10 | CREB1 | 1283 | 5.654 | 0.0707 | No | ||
| 11 | ARNTL | 1310 | 5.592 | 0.0802 | No | ||
| 12 | MGST3 | 1353 | 5.500 | 0.0888 | No | ||
| 13 | HDAC7A | 1425 | 5.354 | 0.0959 | No | ||
| 14 | FBXW11 | 1563 | 5.105 | 0.0994 | No | ||
| 15 | NFAT5 | 1636 | 4.993 | 0.1057 | No | ||
| 16 | MYLK | 1683 | 4.921 | 0.1130 | No | ||
| 17 | ALS2CR2 | 1735 | 4.849 | 0.1200 | No | ||
| 18 | TEF | 1791 | 4.765 | 0.1266 | No | ||
| 19 | CDC42EP3 | 1880 | 4.634 | 0.1314 | No | ||
| 20 | NR4A1 | 2240 | 4.181 | 0.1230 | No | ||
| 21 | LZTS2 | 2258 | 4.161 | 0.1302 | No | ||
| 22 | TMEPAI | 2276 | 4.142 | 0.1374 | No | ||
| 23 | SLAMF1 | 2349 | 4.066 | 0.1419 | No | ||
| 24 | PLAGL2 | 2501 | 3.889 | 0.1424 | No | ||
| 25 | JUN | 2591 | 3.817 | 0.1456 | No | ||
| 26 | CASQ1 | 2600 | 3.808 | 0.1526 | No | ||
| 27 | GNB4 | 2728 | 3.674 | 0.1538 | No | ||
| 28 | NCOR1 | 2965 | 3.464 | 0.1496 | No | ||
| 29 | PURA | 3107 | 3.339 | 0.1496 | No | ||
| 30 | BCL9L | 3239 | 3.231 | 0.1498 | No | ||
| 31 | SMARCA2 | 3274 | 3.202 | 0.1543 | No | ||
| 32 | NEK2 | 3436 | 3.076 | 0.1529 | No | ||
| 33 | PPARGC1A | 3592 | 2.953 | 0.1514 | No | ||
| 34 | PDE6D | 3847 | 2.767 | 0.1451 | No | ||
| 35 | FBXO11 | 4085 | 2.599 | 0.1392 | No | ||
| 36 | CPEB4 | 4224 | 2.491 | 0.1376 | No | ||
| 37 | AMPD1 | 4265 | 2.463 | 0.1405 | No | ||
| 38 | HCFC1R1 | 4387 | 2.393 | 0.1395 | No | ||
| 39 | PTPN1 | 4451 | 2.352 | 0.1412 | No | ||
| 40 | LUZP1 | 4573 | 2.277 | 0.1400 | No | ||
| 41 | ATP1B2 | 4623 | 2.244 | 0.1420 | No | ||
| 42 | AQP1 | 4631 | 2.239 | 0.1460 | No | ||
| 43 | DYRK1B | 4666 | 2.220 | 0.1487 | No | ||
| 44 | ARHGAP26 | 4808 | 2.142 | 0.1464 | No | ||
| 45 | PAK6 | 4864 | 2.116 | 0.1479 | No | ||
| 46 | PHKA2 | 4878 | 2.109 | 0.1513 | No | ||
| 47 | MLLT3 | 5196 | 1.931 | 0.1405 | No | ||
| 48 | GRIN2B | 5286 | 1.882 | 0.1400 | No | ||
| 49 | KCNQ5 | 5382 | 1.841 | 0.1392 | No | ||
| 50 | AAK1 | 5713 | 1.679 | 0.1273 | No | ||
| 51 | OGN | 5771 | 1.654 | 0.1278 | No | ||
| 52 | RG9MTD2 | 6405 | 1.359 | 0.1014 | No | ||
| 53 | WFDC1 | 6591 | 1.263 | 0.0953 | No | ||
| 54 | CASQ2 | 6661 | 1.228 | 0.0945 | No | ||
| 55 | FOS | 6777 | 1.183 | 0.0915 | No | ||
| 56 | ELK4 | 6913 | 1.116 | 0.0874 | No | ||
| 57 | ITGB1BP2 | 7262 | 0.966 | 0.0733 | No | ||
| 58 | HPGD | 7347 | 0.930 | 0.0712 | No | ||
| 59 | BNC2 | 7370 | 0.923 | 0.0720 | No | ||
| 60 | PRDM1 | 7584 | 0.833 | 0.0638 | No | ||
| 61 | CPT1B | 7841 | 0.721 | 0.0534 | No | ||
| 62 | PPP1R3D | 7966 | 0.675 | 0.0490 | No | ||
| 63 | SLC26A6 | 8130 | 0.606 | 0.0427 | No | ||
| 64 | AMMECR1 | 8289 | 0.543 | 0.0365 | No | ||
| 65 | SSPN | 8292 | 0.542 | 0.0374 | No | ||
| 66 | CMYA1 | 8378 | 0.506 | 0.0345 | No | ||
| 67 | MEOX2 | 8437 | 0.480 | 0.0328 | No | ||
| 68 | SMPX | 8545 | 0.434 | 0.0287 | No | ||
| 69 | MYOZ2 | 8966 | 0.249 | 0.0099 | No | ||
| 70 | HLX1 | 9160 | 0.166 | 0.0013 | No | ||
| 71 | MUSK | 9181 | 0.160 | 0.0007 | No | ||
| 72 | PPP2R3A | 9371 | 0.071 | -0.0078 | No | ||
| 73 | EFHD1 | 9584 | -0.033 | -0.0175 | No | ||
| 74 | SMARCA1 | 9698 | -0.084 | -0.0225 | No | ||
| 75 | PPP1R3A | 9833 | -0.151 | -0.0284 | No | ||
| 76 | RALY | 10117 | -0.266 | -0.0409 | No | ||
| 77 | HOXA11 | 10259 | -0.319 | -0.0467 | No | ||
| 78 | IGJ | 10759 | -0.499 | -0.0687 | No | ||
| 79 | GABRB2 | 11000 | -0.580 | -0.0786 | No | ||
| 80 | ADCY2 | 11164 | -0.644 | -0.0849 | No | ||
| 81 | FGF16 | 11306 | -0.683 | -0.0900 | No | ||
| 82 | ANKMY2 | 11350 | -0.696 | -0.0907 | No | ||
| 83 | RAP2C | 12125 | -0.957 | -0.1244 | No | ||
| 84 | NDRG2 | 12273 | -1.000 | -0.1292 | No | ||
| 85 | MGAT3 | 12518 | -1.072 | -0.1384 | No | ||
| 86 | LRRTM4 | 12556 | -1.084 | -0.1380 | No | ||
| 87 | ADAM11 | 12585 | -1.091 | -0.1372 | No | ||
| 88 | SLC8A3 | 12992 | -1.215 | -0.1535 | No | ||
| 89 | IRS1 | 13166 | -1.265 | -0.1590 | No | ||
| 90 | GPC4 | 13223 | -1.281 | -0.1591 | No | ||
| 91 | NBEA | 13402 | -1.333 | -0.1647 | No | ||
| 92 | HFE2 | 13710 | -1.430 | -0.1761 | No | ||
| 93 | RASGEF1B | 13738 | -1.437 | -0.1746 | No | ||
| 94 | TYRO3 | 14002 | -1.507 | -0.1837 | No | ||
| 95 | TCEA3 | 14208 | -1.570 | -0.1901 | No | ||
| 96 | SV2A | 14689 | -1.708 | -0.2089 | No | ||
| 97 | ASB16 | 14703 | -1.713 | -0.2062 | No | ||
| 98 | TNNC2 | 14714 | -1.715 | -0.2034 | No | ||
| 99 | RTBDN | 14869 | -1.761 | -0.2071 | No | ||
| 100 | TRHR | 14898 | -1.771 | -0.2049 | No | ||
| 101 | PTPRO | 15085 | -1.829 | -0.2100 | No | ||
| 102 | CLCN1 | 15203 | -1.866 | -0.2118 | No | ||
| 103 | RASGRP3 | 15577 | -1.992 | -0.2251 | No | ||
| 104 | CD24 | 15636 | -2.010 | -0.2239 | No | ||
| 105 | HSPB3 | 15694 | -2.030 | -0.2226 | No | ||
| 106 | ARHGEF15 | 15857 | -2.081 | -0.2260 | No | ||
| 107 | KCNJ9 | 15858 | -2.082 | -0.2220 | No | ||
| 108 | SLC6A13 | 15930 | -2.102 | -0.2213 | No | ||
| 109 | ESR1 | 16252 | -2.203 | -0.2318 | No | ||
| 110 | HAPLN1 | 16441 | -2.267 | -0.2361 | No | ||
| 111 | MYL1 | 16742 | -2.372 | -0.2453 | Yes | ||
| 112 | SLC30A1 | 16787 | -2.388 | -0.2427 | Yes | ||
| 113 | DMD | 16796 | -2.392 | -0.2385 | Yes | ||
| 114 | FGF12 | 16906 | -2.433 | -0.2388 | Yes | ||
| 115 | ITGA7 | 16908 | -2.433 | -0.2342 | Yes | ||
| 116 | COL8A1 | 17137 | -2.511 | -0.2398 | Yes | ||
| 117 | MEF2C | 17220 | -2.542 | -0.2387 | Yes | ||
| 118 | OPN3 | 17347 | -2.595 | -0.2395 | Yes | ||
| 119 | CSPG3 | 17348 | -2.597 | -0.2345 | Yes | ||
| 120 | USP2 | 17454 | -2.632 | -0.2343 | Yes | ||
| 121 | NR2F1 | 17563 | -2.666 | -0.2342 | Yes | ||
| 122 | SLC2A4 | 17567 | -2.667 | -0.2292 | Yes | ||
| 123 | LRRC20 | 17743 | -2.746 | -0.2319 | Yes | ||
| 124 | EPHA7 | 17815 | -2.779 | -0.2299 | Yes | ||
| 125 | TNNI3K | 17847 | -2.789 | -0.2259 | Yes | ||
| 126 | FOXP2 | 17925 | -2.821 | -0.2240 | Yes | ||
| 127 | CLDN14 | 18035 | -2.870 | -0.2235 | Yes | ||
| 128 | CNTN1 | 18087 | -2.892 | -0.2203 | Yes | ||
| 129 | THBS2 | 18184 | -2.930 | -0.2191 | Yes | ||
| 130 | AP3M1 | 18197 | -2.934 | -0.2140 | Yes | ||
| 131 | HDAC9 | 18201 | -2.936 | -0.2085 | Yes | ||
| 132 | PRRX1 | 18256 | -2.957 | -0.2053 | Yes | ||
| 133 | CDK8 | 18511 | -3.079 | -0.2111 | Yes | ||
| 134 | POU4F1 | 18516 | -3.081 | -0.2053 | Yes | ||
| 135 | ATF3 | 18527 | -3.087 | -0.1999 | Yes | ||
| 136 | MBNL2 | 18541 | -3.091 | -0.1945 | Yes | ||
| 137 | TPP2 | 18549 | -3.093 | -0.1889 | Yes | ||
| 138 | TWIST1 | 18561 | -3.099 | -0.1835 | Yes | ||
| 139 | TRDN | 18571 | -3.102 | -0.1779 | Yes | ||
| 140 | PYY | 18885 | -3.286 | -0.1860 | Yes | ||
| 141 | STAC | 18953 | -3.327 | -0.1827 | Yes | ||
| 142 | RFX4 | 19091 | -3.410 | -0.1824 | Yes | ||
| 143 | RASGRF1 | 19135 | -3.440 | -0.1778 | Yes | ||
| 144 | MFGE8 | 19208 | -3.483 | -0.1744 | Yes | ||
| 145 | ADK | 19431 | -3.635 | -0.1776 | Yes | ||
| 146 | SOX5 | 19470 | -3.663 | -0.1723 | Yes | ||
| 147 | DGKI | 19471 | -3.663 | -0.1653 | Yes | ||
| 148 | DIRAS1 | 19507 | -3.694 | -0.1598 | Yes | ||
| 149 | PIK3R3 | 19528 | -3.708 | -0.1536 | Yes | ||
| 150 | CASK | 19642 | -3.801 | -0.1515 | Yes | ||
| 151 | LBX1 | 19742 | -3.891 | -0.1486 | Yes | ||
| 152 | KCNN1 | 19743 | -3.892 | -0.1411 | Yes | ||
| 153 | GPR85 | 19754 | -3.901 | -0.1341 | Yes | ||
| 154 | ELAVL4 | 19973 | -4.101 | -0.1362 | Yes | ||
| 155 | CTNND2 | 20052 | -4.197 | -0.1317 | Yes | ||
| 156 | SYNPO2L | 20138 | -4.281 | -0.1274 | Yes | ||
| 157 | EYA1 | 20145 | -4.286 | -0.1195 | Yes | ||
| 158 | TPM2 | 20185 | -4.335 | -0.1129 | Yes | ||
| 159 | ART5 | 20215 | -4.372 | -0.1059 | Yes | ||
| 160 | ATP1A2 | 20293 | -4.439 | -0.1009 | Yes | ||
| 161 | EXTL1 | 20324 | -4.478 | -0.0937 | Yes | ||
| 162 | BRUNOL4 | 20358 | -4.522 | -0.0865 | Yes | ||
| 163 | ENO3 | 20436 | -4.643 | -0.0811 | Yes | ||
| 164 | RHOB | 20494 | -4.715 | -0.0747 | Yes | ||
| 165 | KPNA3 | 20663 | -4.935 | -0.0729 | Yes | ||
| 166 | NFIB | 20706 | -5.034 | -0.0652 | Yes | ||
| 167 | TMEM4 | 20785 | -5.175 | -0.0588 | Yes | ||
| 168 | MAB21L2 | 20881 | -5.298 | -0.0530 | Yes | ||
| 169 | HOXA3 | 21069 | -5.643 | -0.0508 | Yes | ||
| 170 | CRTAP | 21546 | -7.110 | -0.0590 | Yes | ||
| 171 | KTN1 | 21583 | -7.244 | -0.0467 | Yes | ||
| 172 | SIPA1L1 | 21621 | -7.482 | -0.0340 | Yes | ||
| 173 | BZW2 | 21862 | -10.683 | -0.0245 | Yes | ||
| 174 | TIMP2 | 21931 | -14.789 | 0.0007 | Yes |
