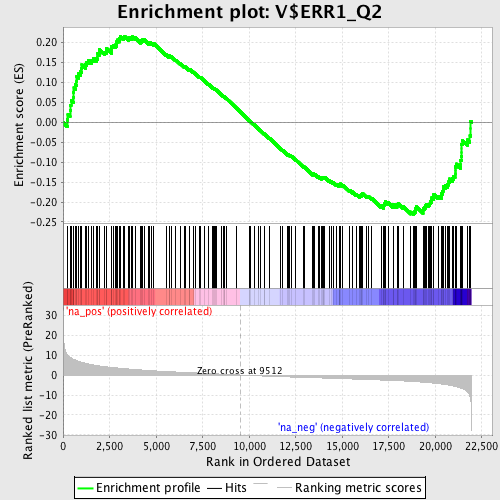
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$ERR1_Q2 |
| Enrichment Score (ES) | -0.2313126 |
| Normalized Enrichment Score (NES) | -1.1488211 |
| Nominal p-value | 0.13017751 |
| FDR q-value | 0.57346004 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ARF6 | 219 | 10.345 | 0.0066 | No | ||
| 2 | SLC37A2 | 260 | 9.832 | 0.0207 | No | ||
| 3 | MAP4K2 | 372 | 8.936 | 0.0300 | No | ||
| 4 | SATB1 | 397 | 8.834 | 0.0432 | No | ||
| 5 | KCNH2 | 427 | 8.532 | 0.0556 | No | ||
| 6 | RNF44 | 564 | 7.862 | 0.0621 | No | ||
| 7 | MSI2 | 575 | 7.805 | 0.0742 | No | ||
| 8 | SMARCC2 | 583 | 7.744 | 0.0864 | No | ||
| 9 | RASGRP2 | 668 | 7.421 | 0.0945 | No | ||
| 10 | CSNK2A1 | 727 | 7.148 | 0.1034 | No | ||
| 11 | ABCC5 | 738 | 7.104 | 0.1144 | No | ||
| 12 | SEMA7A | 817 | 6.836 | 0.1219 | No | ||
| 13 | FBS1 | 937 | 6.504 | 0.1269 | No | ||
| 14 | ID3 | 981 | 6.380 | 0.1352 | No | ||
| 15 | TCF7 | 1011 | 6.319 | 0.1441 | No | ||
| 16 | VASP | 1190 | 5.873 | 0.1454 | No | ||
| 17 | VAMP1 | 1270 | 5.696 | 0.1510 | No | ||
| 18 | MGST3 | 1353 | 5.500 | 0.1561 | No | ||
| 19 | JMJD1C | 1528 | 5.156 | 0.1564 | No | ||
| 20 | IBRDC3 | 1629 | 5.005 | 0.1599 | No | ||
| 21 | TEF | 1791 | 4.765 | 0.1602 | No | ||
| 22 | PSCD2 | 1860 | 4.668 | 0.1646 | No | ||
| 23 | PHF6 | 1864 | 4.663 | 0.1720 | No | ||
| 24 | MYST3 | 1939 | 4.555 | 0.1759 | No | ||
| 25 | ATP5J2 | 1949 | 4.541 | 0.1829 | No | ||
| 26 | NR4A1 | 2240 | 4.181 | 0.1763 | No | ||
| 27 | WBSCR1 | 2315 | 4.098 | 0.1795 | No | ||
| 28 | ACTR1A | 2324 | 4.087 | 0.1857 | No | ||
| 29 | CHD2 | 2595 | 3.816 | 0.1795 | No | ||
| 30 | VAMP2 | 2601 | 3.807 | 0.1854 | No | ||
| 31 | ASCC1 | 2612 | 3.802 | 0.1911 | No | ||
| 32 | PANK1 | 2708 | 3.699 | 0.1927 | No | ||
| 33 | XYLT2 | 2801 | 3.608 | 0.1943 | No | ||
| 34 | SRPK2 | 2883 | 3.531 | 0.1963 | No | ||
| 35 | AFP | 2884 | 3.530 | 0.2020 | No | ||
| 36 | UHRF2 | 2903 | 3.513 | 0.2068 | No | ||
| 37 | JARID1A | 3017 | 3.426 | 0.2072 | No | ||
| 38 | ESRRA | 3039 | 3.404 | 0.2117 | No | ||
| 39 | NR2C2 | 3056 | 3.391 | 0.2164 | No | ||
| 40 | RNF139 | 3269 | 3.211 | 0.2119 | No | ||
| 41 | RAB35 | 3312 | 3.169 | 0.2151 | No | ||
| 42 | SMPD1 | 3516 | 3.008 | 0.2106 | No | ||
| 43 | KIF3B | 3555 | 2.976 | 0.2137 | No | ||
| 44 | ADCY1 | 3687 | 2.884 | 0.2123 | No | ||
| 45 | FRAG1 | 3718 | 2.863 | 0.2155 | No | ||
| 46 | MAP1A | 3878 | 2.742 | 0.2127 | No | ||
| 47 | FBXL17 | 4181 | 2.523 | 0.2029 | No | ||
| 48 | BUB1B | 4223 | 2.491 | 0.2050 | No | ||
| 49 | ATP6V1A | 4266 | 2.462 | 0.2070 | No | ||
| 50 | SART3 | 4346 | 2.413 | 0.2073 | No | ||
| 51 | ATP5A1 | 4601 | 2.260 | 0.1993 | No | ||
| 52 | ITGA3 | 4630 | 2.239 | 0.2016 | No | ||
| 53 | EPN3 | 4767 | 2.161 | 0.1989 | No | ||
| 54 | HTATSF1 | 4881 | 2.107 | 0.1971 | No | ||
| 55 | PRKG2 | 5536 | 1.761 | 0.1699 | No | ||
| 56 | HCRTR1 | 5691 | 1.688 | 0.1655 | No | ||
| 57 | PPM1E | 5694 | 1.688 | 0.1681 | No | ||
| 58 | RFX5 | 5807 | 1.633 | 0.1656 | No | ||
| 59 | RIMS4 | 6061 | 1.514 | 0.1564 | No | ||
| 60 | BCKDHB | 6302 | 1.404 | 0.1477 | No | ||
| 61 | MNT | 6512 | 1.303 | 0.1402 | No | ||
| 62 | USP8 | 6578 | 1.271 | 0.1392 | No | ||
| 63 | RHCG | 6773 | 1.185 | 0.1322 | No | ||
| 64 | SCNN1A | 6781 | 1.180 | 0.1338 | No | ||
| 65 | SPAG9 | 6982 | 1.088 | 0.1264 | No | ||
| 66 | PCDH21 | 7100 | 1.037 | 0.1227 | No | ||
| 67 | PVALB | 7328 | 0.937 | 0.1138 | No | ||
| 68 | KCNG4 | 7367 | 0.924 | 0.1135 | No | ||
| 69 | IDH3G | 7601 | 0.829 | 0.1042 | No | ||
| 70 | RAB3A | 7806 | 0.737 | 0.0960 | No | ||
| 71 | UQCRC1 | 8034 | 0.647 | 0.0866 | No | ||
| 72 | PIM1 | 8068 | 0.635 | 0.0861 | No | ||
| 73 | ZRANB1 | 8131 | 0.604 | 0.0842 | No | ||
| 74 | SNTG1 | 8189 | 0.582 | 0.0825 | No | ||
| 75 | LMO3 | 8247 | 0.558 | 0.0808 | No | ||
| 76 | NXPH4 | 8515 | 0.446 | 0.0693 | No | ||
| 77 | STRN | 8602 | 0.408 | 0.0660 | No | ||
| 78 | FBXL19 | 8665 | 0.384 | 0.0637 | No | ||
| 79 | CLPTM1 | 8675 | 0.379 | 0.0639 | No | ||
| 80 | KIF5A | 8767 | 0.333 | 0.0603 | No | ||
| 81 | STAC2 | 9317 | 0.099 | 0.0352 | No | ||
| 82 | GEMIN7 | 9992 | -0.218 | 0.0046 | No | ||
| 83 | UCHL1 | 9999 | -0.220 | 0.0047 | No | ||
| 84 | GRIK3 | 10059 | -0.246 | 0.0024 | No | ||
| 85 | FZD9 | 10258 | -0.319 | -0.0062 | No | ||
| 86 | HDAC11 | 10521 | -0.412 | -0.0176 | No | ||
| 87 | ABAT | 10584 | -0.433 | -0.0197 | No | ||
| 88 | SLC30A3 | 10803 | -0.513 | -0.0289 | No | ||
| 89 | LHX2 | 10846 | -0.527 | -0.0300 | No | ||
| 90 | SNAG1 | 11077 | -0.608 | -0.0396 | No | ||
| 91 | DLC1 | 11686 | -0.820 | -0.0662 | No | ||
| 92 | CDH16 | 11811 | -0.860 | -0.0705 | No | ||
| 93 | RGS4 | 12075 | -0.942 | -0.0811 | No | ||
| 94 | RAP2C | 12125 | -0.957 | -0.0818 | No | ||
| 95 | HOXA5 | 12187 | -0.972 | -0.0830 | No | ||
| 96 | NDRG2 | 12273 | -1.000 | -0.0853 | No | ||
| 97 | ZDHHC21 | 12495 | -1.067 | -0.0937 | No | ||
| 98 | SOD3 | 12899 | -1.186 | -0.1103 | No | ||
| 99 | FGF9 | 12977 | -1.209 | -0.1119 | No | ||
| 100 | NEF3 | 13424 | -1.340 | -0.1303 | No | ||
| 101 | PDCD8 | 13450 | -1.347 | -0.1292 | No | ||
| 102 | NOG | 13521 | -1.371 | -0.1302 | No | ||
| 103 | SDHB | 13698 | -1.428 | -0.1360 | No | ||
| 104 | SCN5A | 13788 | -1.447 | -0.1378 | No | ||
| 105 | CACNB2 | 13894 | -1.477 | -0.1402 | No | ||
| 106 | OBSCN | 13924 | -1.485 | -0.1391 | No | ||
| 107 | ANKRD28 | 13973 | -1.498 | -0.1389 | No | ||
| 108 | RYR3 | 13994 | -1.504 | -0.1374 | No | ||
| 109 | NTF3 | 14066 | -1.527 | -0.1382 | No | ||
| 110 | SLC38A3 | 14310 | -1.600 | -0.1468 | No | ||
| 111 | PLCB3 | 14401 | -1.621 | -0.1483 | No | ||
| 112 | ELAVL3 | 14529 | -1.656 | -0.1515 | No | ||
| 113 | PPP2R5D | 14684 | -1.704 | -0.1558 | No | ||
| 114 | TGM6 | 14826 | -1.749 | -0.1595 | No | ||
| 115 | TCF2 | 14838 | -1.752 | -0.1571 | No | ||
| 116 | LRFN4 | 14861 | -1.758 | -0.1553 | No | ||
| 117 | MMP24 | 14906 | -1.772 | -0.1545 | No | ||
| 118 | SAG | 15038 | -1.809 | -0.1576 | No | ||
| 119 | TFEB | 15407 | -1.927 | -0.1714 | No | ||
| 120 | SCNN1G | 15547 | -1.980 | -0.1746 | No | ||
| 121 | MB | 15750 | -2.046 | -0.1805 | No | ||
| 122 | PSIP1 | 15945 | -2.106 | -0.1861 | No | ||
| 123 | HSPE1 | 15969 | -2.115 | -0.1837 | No | ||
| 124 | CNTN2 | 16016 | -2.126 | -0.1824 | No | ||
| 125 | MYL3 | 16041 | -2.134 | -0.1800 | No | ||
| 126 | HECTD2 | 16080 | -2.146 | -0.1783 | No | ||
| 127 | SLC31A2 | 16327 | -2.224 | -0.1860 | No | ||
| 128 | SPATS1 | 16391 | -2.245 | -0.1853 | No | ||
| 129 | TBC1D15 | 16586 | -2.320 | -0.1905 | No | ||
| 130 | PCDH7 | 17094 | -2.497 | -0.2097 | No | ||
| 131 | MEF2C | 17220 | -2.542 | -0.2114 | No | ||
| 132 | CX3CL1 | 17229 | -2.546 | -0.2076 | No | ||
| 133 | RPH3A | 17250 | -2.554 | -0.2044 | No | ||
| 134 | SSR4 | 17297 | -2.573 | -0.2024 | No | ||
| 135 | NAV1 | 17308 | -2.577 | -0.1987 | No | ||
| 136 | RAPGEF5 | 17471 | -2.638 | -0.2019 | No | ||
| 137 | HS6ST2 | 17746 | -2.747 | -0.2100 | No | ||
| 138 | VSNL1 | 17748 | -2.748 | -0.2056 | No | ||
| 139 | ATP6AP2 | 17941 | -2.830 | -0.2099 | No | ||
| 140 | KCNK1 | 17947 | -2.832 | -0.2055 | No | ||
| 141 | PHF15 | 18005 | -2.856 | -0.2035 | No | ||
| 142 | SORCS1 | 18281 | -2.967 | -0.2114 | No | ||
| 143 | RASAL1 | 18690 | -3.173 | -0.2250 | No | ||
| 144 | LDB3 | 18828 | -3.252 | -0.2261 | Yes | ||
| 145 | RNF128 | 18899 | -3.297 | -0.2240 | Yes | ||
| 146 | FGL1 | 18947 | -3.324 | -0.2207 | Yes | ||
| 147 | UNC5B | 18949 | -3.324 | -0.2154 | Yes | ||
| 148 | TMEM28 | 18991 | -3.349 | -0.2119 | Yes | ||
| 149 | YTHDF2 | 19339 | -3.567 | -0.2221 | Yes | ||
| 150 | THRA | 19344 | -3.570 | -0.2165 | Yes | ||
| 151 | CXXC5 | 19406 | -3.615 | -0.2135 | Yes | ||
| 152 | KIRREL3 | 19452 | -3.649 | -0.2096 | Yes | ||
| 153 | DIRAS1 | 19507 | -3.694 | -0.2062 | Yes | ||
| 154 | GJB2 | 19637 | -3.796 | -0.2060 | Yes | ||
| 155 | LIMK1 | 19712 | -3.863 | -0.2031 | Yes | ||
| 156 | KCNN1 | 19743 | -3.892 | -0.1982 | Yes | ||
| 157 | WDFY3 | 19807 | -3.956 | -0.1947 | Yes | ||
| 158 | SEC61A1 | 19817 | -3.963 | -0.1887 | Yes | ||
| 159 | ACO2 | 19914 | -4.047 | -0.1866 | Yes | ||
| 160 | RNF2 | 19919 | -4.053 | -0.1802 | Yes | ||
| 161 | ANGPT1 | 20181 | -4.330 | -0.1852 | Yes | ||
| 162 | UQCRC2 | 20319 | -4.470 | -0.1843 | Yes | ||
| 163 | ADAM9 | 20336 | -4.490 | -0.1778 | Yes | ||
| 164 | MRPL48 | 20390 | -4.564 | -0.1729 | Yes | ||
| 165 | SLC25A5 | 20419 | -4.616 | -0.1667 | Yes | ||
| 166 | ENO3 | 20436 | -4.643 | -0.1599 | Yes | ||
| 167 | AFG3L2 | 20555 | -4.780 | -0.1577 | Yes | ||
| 168 | APP | 20670 | -4.957 | -0.1549 | Yes | ||
| 169 | PHF5A | 20728 | -5.077 | -0.1493 | Yes | ||
| 170 | JARID1C | 20752 | -5.114 | -0.1421 | Yes | ||
| 171 | HSPD1 | 20908 | -5.331 | -0.1406 | Yes | ||
| 172 | RREB1 | 20991 | -5.466 | -0.1356 | Yes | ||
| 173 | VDAC2 | 21060 | -5.633 | -0.1296 | Yes | ||
| 174 | VDAC1 | 21066 | -5.638 | -0.1207 | Yes | ||
| 175 | HOXA3 | 21069 | -5.643 | -0.1117 | Yes | ||
| 176 | SHB | 21116 | -5.744 | -0.1045 | Yes | ||
| 177 | SOCS4 | 21347 | -6.426 | -0.1047 | Yes | ||
| 178 | SUCLA2 | 21355 | -6.441 | -0.0946 | Yes | ||
| 179 | SDHD | 21390 | -6.548 | -0.0856 | Yes | ||
| 180 | ATP5G1 | 21410 | -6.613 | -0.0758 | Yes | ||
| 181 | VCL | 21412 | -6.617 | -0.0652 | Yes | ||
| 182 | TIMM8B | 21413 | -6.619 | -0.0545 | Yes | ||
| 183 | PHB | 21449 | -6.759 | -0.0452 | Yes | ||
| 184 | TIMM9 | 21713 | -8.175 | -0.0441 | Yes | ||
| 185 | BZW2 | 21862 | -10.683 | -0.0336 | Yes | ||
| 186 | GOT1 | 21887 | -11.570 | -0.0160 | Yes | ||
| 187 | PCBP4 | 21889 | -11.615 | 0.0027 | Yes |
