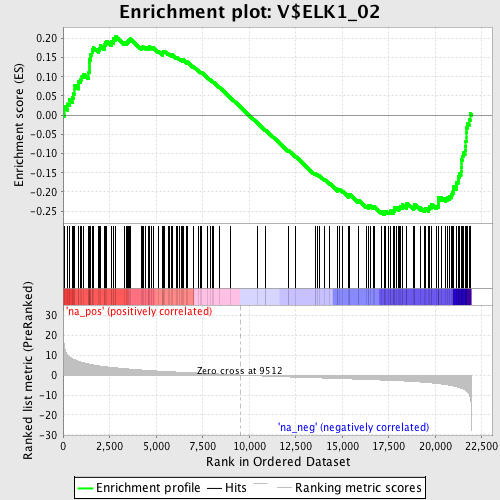
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$ELK1_02 |
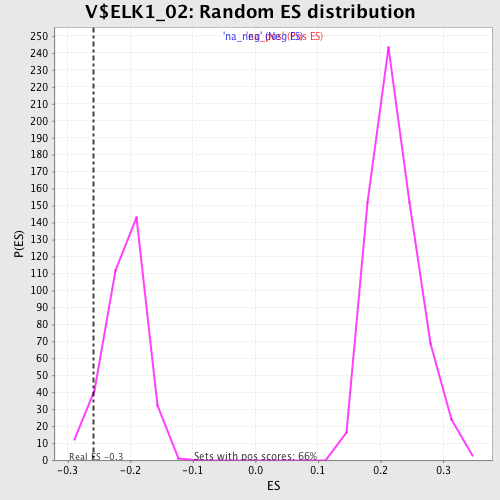
| Enrichment Score (ES) | -0.25871724 |
| Normalized Enrichment Score (NES) | -1.2406073 |
| Nominal p-value | 0.07038123 |
| FDR q-value | 0.44261387 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BCL2L1 | 52 | 15.577 | 0.0224 | No | ||
| 2 | SENP7 | 255 | 9.945 | 0.0290 | No | ||
| 3 | TBCC | 339 | 9.185 | 0.0398 | No | ||
| 4 | ACTR2 | 500 | 8.150 | 0.0455 | No | ||
| 5 | AP2S1 | 557 | 7.886 | 0.0554 | No | ||
| 6 | DHX30 | 590 | 7.724 | 0.0663 | No | ||
| 7 | NR1H2 | 595 | 7.693 | 0.0784 | No | ||
| 8 | RAB11B | 821 | 6.811 | 0.0789 | No | ||
| 9 | USP32 | 844 | 6.751 | 0.0886 | No | ||
| 10 | DNAJC7 | 951 | 6.459 | 0.0940 | No | ||
| 11 | TCF7 | 1011 | 6.319 | 0.1014 | No | ||
| 12 | HERC4 | 1104 | 6.077 | 0.1069 | No | ||
| 13 | INSIG2 | 1383 | 5.424 | 0.1027 | No | ||
| 14 | GTF3C1 | 1384 | 5.424 | 0.1114 | No | ||
| 15 | AP1M1 | 1406 | 5.387 | 0.1190 | No | ||
| 16 | MYST2 | 1409 | 5.380 | 0.1275 | No | ||
| 17 | SIRT6 | 1413 | 5.374 | 0.1359 | No | ||
| 18 | PCQAP | 1419 | 5.363 | 0.1442 | No | ||
| 19 | NUP214 | 1449 | 5.310 | 0.1513 | No | ||
| 20 | UBADC1 | 1468 | 5.270 | 0.1589 | No | ||
| 21 | GABARAPL2 | 1552 | 5.124 | 0.1633 | No | ||
| 22 | SUPT5H | 1568 | 5.096 | 0.1707 | No | ||
| 23 | SEC24C | 1616 | 5.021 | 0.1765 | No | ||
| 24 | DNAJA2 | 1916 | 4.577 | 0.1701 | No | ||
| 25 | LMAN2 | 1975 | 4.498 | 0.1746 | No | ||
| 26 | SNRPB | 1987 | 4.478 | 0.1812 | No | ||
| 27 | TRAF7 | 2206 | 4.221 | 0.1779 | No | ||
| 28 | MAPKAP1 | 2213 | 4.210 | 0.1844 | No | ||
| 29 | HMG20A | 2266 | 4.152 | 0.1886 | No | ||
| 30 | TBC1D17 | 2343 | 4.074 | 0.1916 | No | ||
| 31 | PRKACB | 2592 | 3.816 | 0.1863 | No | ||
| 32 | RAPH1 | 2594 | 3.816 | 0.1923 | No | ||
| 33 | LEPRE1 | 2705 | 3.702 | 0.1932 | No | ||
| 34 | SHC1 | 2716 | 3.694 | 0.1986 | No | ||
| 35 | STK4 | 2809 | 3.603 | 0.2001 | No | ||
| 36 | GOLGA1 | 2812 | 3.600 | 0.2057 | No | ||
| 37 | ARHGAP1 | 3297 | 3.177 | 0.1886 | No | ||
| 38 | TBX6 | 3402 | 3.103 | 0.1887 | No | ||
| 39 | GTF3C2 | 3452 | 3.062 | 0.1914 | No | ||
| 40 | IL2RG | 3507 | 3.016 | 0.1937 | No | ||
| 41 | PURB | 3560 | 2.973 | 0.1960 | No | ||
| 42 | MARK3 | 3610 | 2.938 | 0.1985 | No | ||
| 43 | CPEB4 | 4224 | 2.491 | 0.1743 | No | ||
| 44 | UBR1 | 4241 | 2.476 | 0.1775 | No | ||
| 45 | ARCN1 | 4328 | 2.421 | 0.1774 | No | ||
| 46 | PCYT1A | 4445 | 2.355 | 0.1758 | No | ||
| 47 | AP1B1 | 4572 | 2.277 | 0.1737 | No | ||
| 48 | ATP5A1 | 4601 | 2.260 | 0.1760 | No | ||
| 49 | CHERP | 4624 | 2.242 | 0.1786 | No | ||
| 50 | BTAF1 | 4764 | 2.163 | 0.1756 | No | ||
| 51 | ITGB3BP | 4840 | 2.129 | 0.1756 | No | ||
| 52 | CDC45L | 5101 | 1.977 | 0.1668 | No | ||
| 53 | DIABLO | 5333 | 1.861 | 0.1591 | No | ||
| 54 | TRIM46 | 5361 | 1.850 | 0.1608 | No | ||
| 55 | SAMD8 | 5367 | 1.849 | 0.1636 | No | ||
| 56 | EPN1 | 5377 | 1.843 | 0.1661 | No | ||
| 57 | ZDHHC5 | 5444 | 1.812 | 0.1659 | No | ||
| 58 | NKIRAS2 | 5648 | 1.702 | 0.1593 | No | ||
| 59 | AAK1 | 5713 | 1.679 | 0.1591 | No | ||
| 60 | SMUG1 | 5846 | 1.618 | 0.1556 | No | ||
| 61 | PRCP | 5853 | 1.615 | 0.1579 | No | ||
| 62 | COG3 | 6085 | 1.508 | 0.1497 | No | ||
| 63 | BMP2K | 6122 | 1.491 | 0.1504 | No | ||
| 64 | ACP2 | 6269 | 1.419 | 0.1460 | No | ||
| 65 | NRAS | 6347 | 1.384 | 0.1446 | No | ||
| 66 | KRTCAP2 | 6436 | 1.344 | 0.1427 | No | ||
| 67 | LYPLA2 | 6442 | 1.340 | 0.1446 | No | ||
| 68 | FBXO3 | 6643 | 1.235 | 0.1374 | No | ||
| 69 | NEDD1 | 6656 | 1.230 | 0.1388 | No | ||
| 70 | NF1 | 6995 | 1.084 | 0.1250 | No | ||
| 71 | PHKB | 7021 | 1.071 | 0.1256 | No | ||
| 72 | NFYA | 7281 | 0.960 | 0.1152 | No | ||
| 73 | PRPF18 | 7403 | 0.908 | 0.1111 | No | ||
| 74 | RPL19 | 7419 | 0.900 | 0.1119 | No | ||
| 75 | TUFM | 7766 | 0.753 | 0.0972 | No | ||
| 76 | PDZRN4 | 7902 | 0.702 | 0.0921 | No | ||
| 77 | HSPC171 | 8031 | 0.647 | 0.0872 | No | ||
| 78 | SON | 8100 | 0.621 | 0.0851 | No | ||
| 79 | ZBTB11 | 8410 | 0.494 | 0.0717 | No | ||
| 80 | SCO1 | 8980 | 0.244 | 0.0460 | No | ||
| 81 | EEF1B2 | 10449 | -0.388 | -0.0208 | No | ||
| 82 | ZNF22 | 10860 | -0.533 | -0.0388 | No | ||
| 83 | BAT3 | 10896 | -0.547 | -0.0395 | No | ||
| 84 | RPL37 | 12086 | -0.944 | -0.0926 | No | ||
| 85 | RAP2C | 12125 | -0.957 | -0.0928 | No | ||
| 86 | CLDN5 | 12486 | -1.063 | -0.1077 | No | ||
| 87 | AKT1S1 | 13535 | -1.374 | -0.1536 | No | ||
| 88 | PPP2R2C | 13559 | -1.382 | -0.1525 | No | ||
| 89 | EN1 | 13659 | -1.411 | -0.1548 | No | ||
| 90 | ZNF23 | 13753 | -1.439 | -0.1567 | No | ||
| 91 | EIF4G1 | 14061 | -1.526 | -0.1684 | No | ||
| 92 | FBXW9 | 14308 | -1.599 | -0.1772 | No | ||
| 93 | UQCRH | 14758 | -1.729 | -0.1950 | No | ||
| 94 | TRO | 14760 | -1.730 | -0.1923 | No | ||
| 95 | OGG1 | 14843 | -1.753 | -0.1933 | No | ||
| 96 | CALU | 15009 | -1.803 | -0.1980 | No | ||
| 97 | MED8 | 15350 | -1.909 | -0.2106 | No | ||
| 98 | GGTL3 | 15372 | -1.919 | -0.2085 | No | ||
| 99 | FANCD2 | 15380 | -1.921 | -0.2057 | No | ||
| 100 | TSTA3 | 15882 | -2.088 | -0.2254 | No | ||
| 101 | LSM5 | 15885 | -2.089 | -0.2222 | No | ||
| 102 | TRIM44 | 16281 | -2.212 | -0.2368 | No | ||
| 103 | SYT5 | 16410 | -2.253 | -0.2391 | No | ||
| 104 | PPIG | 16414 | -2.255 | -0.2356 | No | ||
| 105 | RAB2 | 16500 | -2.289 | -0.2359 | No | ||
| 106 | SMS | 16654 | -2.338 | -0.2392 | No | ||
| 107 | PPIE | 16708 | -2.359 | -0.2378 | No | ||
| 108 | CLDN7 | 17085 | -2.494 | -0.2511 | No | ||
| 109 | STX12 | 17251 | -2.554 | -0.2546 | Yes | ||
| 110 | ZF | 17282 | -2.566 | -0.2519 | Yes | ||
| 111 | SRP19 | 17326 | -2.585 | -0.2498 | Yes | ||
| 112 | FOXH1 | 17476 | -2.639 | -0.2524 | Yes | ||
| 113 | CRB3 | 17586 | -2.676 | -0.2532 | Yes | ||
| 114 | DDX5 | 17599 | -2.683 | -0.2495 | Yes | ||
| 115 | PRKAG2 | 17729 | -2.739 | -0.2510 | Yes | ||
| 116 | FMR1 | 17747 | -2.747 | -0.2474 | Yes | ||
| 117 | ITM2C | 17808 | -2.776 | -0.2457 | Yes | ||
| 118 | NUDT12 | 17812 | -2.778 | -0.2415 | Yes | ||
| 119 | HAT1 | 17894 | -2.808 | -0.2407 | Yes | ||
| 120 | UGCGL2 | 18040 | -2.872 | -0.2428 | Yes | ||
| 121 | RBM22 | 18084 | -2.892 | -0.2402 | Yes | ||
| 122 | KLHDC3 | 18114 | -2.904 | -0.2369 | Yes | ||
| 123 | HMGA1 | 18236 | -2.950 | -0.2377 | Yes | ||
| 124 | SLC30A7 | 18246 | -2.952 | -0.2334 | Yes | ||
| 125 | MTPN | 18448 | -3.049 | -0.2378 | Yes | ||
| 126 | NDUFS1 | 18468 | -3.057 | -0.2338 | Yes | ||
| 127 | DPP8 | 18471 | -3.060 | -0.2290 | Yes | ||
| 128 | EIF5A | 18853 | -3.265 | -0.2413 | Yes | ||
| 129 | CDC2L5 | 18880 | -3.280 | -0.2373 | Yes | ||
| 130 | FBXO22 | 18893 | -3.291 | -0.2326 | Yes | ||
| 131 | DDOST | 19212 | -3.487 | -0.2416 | Yes | ||
| 132 | STOML2 | 19403 | -3.613 | -0.2446 | Yes | ||
| 133 | PSMC1 | 19490 | -3.677 | -0.2427 | Yes | ||
| 134 | TRMT1 | 19656 | -3.812 | -0.2442 | Yes | ||
| 135 | POLL | 19672 | -3.826 | -0.2388 | Yes | ||
| 136 | NAT5 | 19792 | -3.941 | -0.2380 | Yes | ||
| 137 | MTX2 | 19794 | -3.941 | -0.2317 | Yes | ||
| 138 | UFD1L | 20068 | -4.207 | -0.2376 | Yes | ||
| 139 | ELAVL1 | 20152 | -4.291 | -0.2346 | Yes | ||
| 140 | RFC4 | 20169 | -4.311 | -0.2284 | Yes | ||
| 141 | MAPK8IP3 | 20178 | -4.329 | -0.2219 | Yes | ||
| 142 | COX8A | 20186 | -4.336 | -0.2153 | Yes | ||
| 143 | GTF2A2 | 20323 | -4.477 | -0.2144 | Yes | ||
| 144 | UBE2N | 20532 | -4.758 | -0.2164 | Yes | ||
| 145 | EXTL2 | 20646 | -4.909 | -0.2138 | Yes | ||
| 146 | PPHLN1 | 20761 | -5.129 | -0.2108 | Yes | ||
| 147 | SEC13L1 | 20871 | -5.285 | -0.2074 | Yes | ||
| 148 | PSMD12 | 20898 | -5.320 | -0.2001 | Yes | ||
| 149 | MTBP | 20975 | -5.438 | -0.1950 | Yes | ||
| 150 | FBXO36 | 20983 | -5.456 | -0.1866 | Yes | ||
| 151 | IPO4 | 21131 | -5.788 | -0.1841 | Yes | ||
| 152 | UBE2L3 | 21148 | -5.829 | -0.1756 | Yes | ||
| 153 | PAPD4 | 21221 | -6.045 | -0.1692 | Yes | ||
| 154 | RPS18 | 21243 | -6.113 | -0.1605 | Yes | ||
| 155 | CRK | 21299 | -6.288 | -0.1530 | Yes | ||
| 156 | CGGBP1 | 21380 | -6.513 | -0.1463 | Yes | ||
| 157 | SDHD | 21390 | -6.548 | -0.1362 | Yes | ||
| 158 | PEX11A | 21411 | -6.616 | -0.1266 | Yes | ||
| 159 | TIMM8B | 21413 | -6.619 | -0.1161 | Yes | ||
| 160 | CKS1B | 21442 | -6.729 | -0.1067 | Yes | ||
| 161 | ITGB1BP1 | 21492 | -6.893 | -0.0980 | Yes | ||
| 162 | IMMT | 21596 | -7.320 | -0.0910 | Yes | ||
| 163 | PSMC2 | 21627 | -7.542 | -0.0804 | Yes | ||
| 164 | MRPL3 | 21642 | -7.624 | -0.0689 | Yes | ||
| 165 | PSMC6 | 21665 | -7.755 | -0.0575 | Yes | ||
| 166 | MIR16 | 21667 | -7.779 | -0.0452 | Yes | ||
| 167 | CPSF3 | 21681 | -7.899 | -0.0332 | Yes | ||
| 168 | TOMM40 | 21709 | -8.157 | -0.0215 | Yes | ||
| 169 | MRPL13 | 21813 | -9.340 | -0.0113 | Yes | ||
| 170 | GART | 21869 | -10.928 | 0.0036 | Yes |