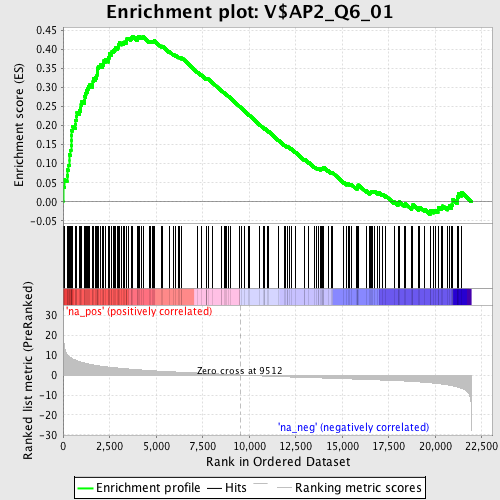
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$AP2_Q6_01 |
| Enrichment Score (ES) | 0.43423843 |
| Normalized Enrichment Score (NES) | 1.9456035 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0063560936 |
| FWER p-Value | 0.023 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EGR3 | 3 | 25.122 | 0.0400 | Yes | ||
| 2 | BCL6 | 77 | 13.547 | 0.0583 | Yes | ||
| 3 | MGEA5 | 212 | 10.405 | 0.0688 | Yes | ||
| 4 | CIC | 237 | 10.141 | 0.0839 | Yes | ||
| 5 | CRY1 | 301 | 9.519 | 0.0962 | Yes | ||
| 6 | PHF1 | 335 | 9.207 | 0.1094 | Yes | ||
| 7 | THRAP1 | 337 | 9.200 | 0.1241 | Yes | ||
| 8 | POFUT1 | 399 | 8.796 | 0.1353 | Yes | ||
| 9 | KCNH2 | 427 | 8.532 | 0.1477 | Yes | ||
| 10 | RNF40 | 439 | 8.481 | 0.1608 | Yes | ||
| 11 | RFX1 | 446 | 8.462 | 0.1740 | Yes | ||
| 12 | MTCH2 | 455 | 8.423 | 0.1871 | Yes | ||
| 13 | ARF3 | 491 | 8.188 | 0.1986 | Yes | ||
| 14 | RBM6 | 652 | 7.471 | 0.2032 | Yes | ||
| 15 | RASGRP2 | 668 | 7.421 | 0.2144 | Yes | ||
| 16 | ATP5H | 722 | 7.172 | 0.2234 | Yes | ||
| 17 | SMAD7 | 742 | 7.089 | 0.2338 | Yes | ||
| 18 | FNBP1 | 866 | 6.684 | 0.2389 | Yes | ||
| 19 | FBS1 | 937 | 6.504 | 0.2460 | Yes | ||
| 20 | BCAT2 | 960 | 6.433 | 0.2553 | Yes | ||
| 21 | NRF1 | 1006 | 6.329 | 0.2634 | Yes | ||
| 22 | NCOA6 | 1139 | 6.005 | 0.2669 | Yes | ||
| 23 | DNMT3A | 1150 | 5.973 | 0.2760 | Yes | ||
| 24 | MKNK2 | 1193 | 5.865 | 0.2834 | Yes | ||
| 25 | SIPA1 | 1231 | 5.786 | 0.2910 | Yes | ||
| 26 | BAHD1 | 1315 | 5.575 | 0.2961 | Yes | ||
| 27 | CAMTA2 | 1345 | 5.516 | 0.3036 | Yes | ||
| 28 | MLL5 | 1438 | 5.327 | 0.3078 | Yes | ||
| 29 | TLK2 | 1596 | 5.060 | 0.3087 | Yes | ||
| 30 | FMNL1 | 1603 | 5.043 | 0.3165 | Yes | ||
| 31 | KIF3C | 1622 | 5.012 | 0.3237 | Yes | ||
| 32 | JMJD1B | 1729 | 4.855 | 0.3266 | Yes | ||
| 33 | CHD4 | 1814 | 4.734 | 0.3303 | Yes | ||
| 34 | TRIM28 | 1824 | 4.714 | 0.3374 | Yes | ||
| 35 | CRSP7 | 1847 | 4.683 | 0.3439 | Yes | ||
| 36 | PSCD2 | 1860 | 4.668 | 0.3508 | Yes | ||
| 37 | B4GALT1 | 1910 | 4.582 | 0.3559 | Yes | ||
| 38 | GRK6 | 1983 | 4.483 | 0.3597 | Yes | ||
| 39 | SLC12A4 | 2139 | 4.291 | 0.3594 | Yes | ||
| 40 | FBXL11 | 2188 | 4.233 | 0.3640 | Yes | ||
| 41 | TERF2 | 2193 | 4.230 | 0.3706 | Yes | ||
| 42 | MESDC1 | 2292 | 4.119 | 0.3727 | Yes | ||
| 43 | PLXNC1 | 2431 | 3.971 | 0.3727 | Yes | ||
| 44 | PCTK1 | 2434 | 3.967 | 0.3789 | Yes | ||
| 45 | PICALM | 2482 | 3.908 | 0.3830 | Yes | ||
| 46 | PLAGL2 | 2501 | 3.889 | 0.3884 | Yes | ||
| 47 | CNP | 2618 | 3.795 | 0.3891 | Yes | ||
| 48 | CYB561D1 | 2623 | 3.788 | 0.3950 | Yes | ||
| 49 | CELSR2 | 2696 | 3.707 | 0.3976 | Yes | ||
| 50 | UBE2S | 2775 | 3.635 | 0.3998 | Yes | ||
| 51 | CENTD2 | 2822 | 3.596 | 0.4035 | Yes | ||
| 52 | BCOR | 2930 | 3.491 | 0.4041 | Yes | ||
| 53 | DMPK | 2956 | 3.470 | 0.4085 | Yes | ||
| 54 | TRIM39 | 2994 | 3.442 | 0.4123 | Yes | ||
| 55 | OAZ2 | 3008 | 3.430 | 0.4172 | Yes | ||
| 56 | CDK9 | 3146 | 3.309 | 0.4162 | Yes | ||
| 57 | LDB1 | 3233 | 3.237 | 0.4174 | Yes | ||
| 58 | RAB35 | 3312 | 3.169 | 0.4189 | Yes | ||
| 59 | SLC37A4 | 3385 | 3.113 | 0.4206 | Yes | ||
| 60 | UBTF | 3415 | 3.094 | 0.4242 | Yes | ||
| 61 | HOXB2 | 3430 | 3.081 | 0.4285 | Yes | ||
| 62 | TACC2 | 3525 | 3.001 | 0.4290 | Yes | ||
| 63 | NFYC | 3647 | 2.909 | 0.4280 | Yes | ||
| 64 | GLTP | 3693 | 2.878 | 0.4306 | Yes | ||
| 65 | KCNH8 | 3714 | 2.865 | 0.4342 | Yes | ||
| 66 | SYMPK | 3978 | 2.657 | 0.4264 | No | ||
| 67 | CNNM4 | 3989 | 2.653 | 0.4302 | No | ||
| 68 | CTCF | 4032 | 2.627 | 0.4325 | No | ||
| 69 | NXPH3 | 4120 | 2.575 | 0.4326 | No | ||
| 70 | SPRED1 | 4231 | 2.484 | 0.4315 | No | ||
| 71 | GPR12 | 4294 | 2.446 | 0.4325 | No | ||
| 72 | VEGF | 4617 | 2.251 | 0.4213 | No | ||
| 73 | RDH10 | 4706 | 2.195 | 0.4208 | No | ||
| 74 | CHAT | 4793 | 2.150 | 0.4203 | No | ||
| 75 | DTNB | 4877 | 2.109 | 0.4199 | No | ||
| 76 | SLC25A28 | 4888 | 2.103 | 0.4228 | No | ||
| 77 | NCOA2 | 5268 | 1.893 | 0.4084 | No | ||
| 78 | RAD21 | 5365 | 1.849 | 0.4069 | No | ||
| 79 | PPM1E | 5694 | 1.688 | 0.3945 | No | ||
| 80 | ADRB1 | 5918 | 1.585 | 0.3868 | No | ||
| 81 | GP1BB | 6051 | 1.520 | 0.3832 | No | ||
| 82 | PIAS1 | 6189 | 1.455 | 0.3792 | No | ||
| 83 | MAP3K7IP2 | 6233 | 1.435 | 0.3795 | No | ||
| 84 | TMEM25 | 6351 | 1.383 | 0.3764 | No | ||
| 85 | POU5F1 | 6381 | 1.370 | 0.3772 | No | ||
| 86 | MAFB | 7229 | 0.984 | 0.3399 | No | ||
| 87 | FASTK | 7422 | 0.899 | 0.3325 | No | ||
| 88 | FXYD1 | 7678 | 0.793 | 0.3220 | No | ||
| 89 | SLC18A3 | 7679 | 0.793 | 0.3233 | No | ||
| 90 | MGAT4B | 7711 | 0.777 | 0.3231 | No | ||
| 91 | HOXA1 | 7728 | 0.769 | 0.3236 | No | ||
| 92 | FOXA3 | 7788 | 0.746 | 0.3221 | No | ||
| 93 | CPSF4 | 8043 | 0.644 | 0.3115 | No | ||
| 94 | ITPKA | 8518 | 0.445 | 0.2904 | No | ||
| 95 | RHBDF1 | 8524 | 0.443 | 0.2909 | No | ||
| 96 | ACCN1 | 8657 | 0.386 | 0.2854 | No | ||
| 97 | GNG8 | 8718 | 0.356 | 0.2832 | No | ||
| 98 | KLF13 | 8804 | 0.321 | 0.2798 | No | ||
| 99 | SLC17A7 | 8882 | 0.290 | 0.2768 | No | ||
| 100 | STMN1 | 8975 | 0.246 | 0.2729 | No | ||
| 101 | IL11 | 9486 | 0.018 | 0.2495 | No | ||
| 102 | KCNJ12 | 9487 | 0.017 | 0.2496 | No | ||
| 103 | GPR3 | 9559 | -0.022 | 0.2463 | No | ||
| 104 | EFNB3 | 9743 | -0.106 | 0.2381 | No | ||
| 105 | SLC7A10 | 9979 | -0.212 | 0.2276 | No | ||
| 106 | ARID1A | 10021 | -0.231 | 0.2261 | No | ||
| 107 | SFRS16 | 10568 | -0.429 | 0.2017 | No | ||
| 108 | ADAMTS5 | 10756 | -0.499 | 0.1939 | No | ||
| 109 | SOX2 | 10843 | -0.526 | 0.1908 | No | ||
| 110 | LHX2 | 10846 | -0.527 | 0.1916 | No | ||
| 111 | PGF | 10994 | -0.579 | 0.1857 | No | ||
| 112 | SLC6A11 | 11059 | -0.599 | 0.1837 | No | ||
| 113 | HOXB9 | 11583 | -0.779 | 0.1610 | No | ||
| 114 | PAQR4 | 11878 | -0.882 | 0.1489 | No | ||
| 115 | PORCN | 11940 | -0.902 | 0.1475 | No | ||
| 116 | CEECAM1 | 12046 | -0.934 | 0.1442 | No | ||
| 117 | SALL1 | 12077 | -0.942 | 0.1443 | No | ||
| 118 | KCNH5 | 12154 | -0.962 | 0.1423 | No | ||
| 119 | ANP32A | 12262 | -0.996 | 0.1390 | No | ||
| 120 | CLDN5 | 12486 | -1.063 | 0.1305 | No | ||
| 121 | HEY1 | 12952 | -1.200 | 0.1110 | No | ||
| 122 | FGF9 | 12977 | -1.209 | 0.1118 | No | ||
| 123 | FGD1 | 13206 | -1.276 | 0.1034 | No | ||
| 124 | NTF5 | 13523 | -1.372 | 0.0911 | No | ||
| 125 | SH3BP4 | 13632 | -1.401 | 0.0884 | No | ||
| 126 | IGSF4 | 13695 | -1.427 | 0.0878 | No | ||
| 127 | PRDM16 | 13728 | -1.434 | 0.0886 | No | ||
| 128 | RASIP1 | 13848 | -1.463 | 0.0855 | No | ||
| 129 | MN1 | 13857 | -1.466 | 0.0875 | No | ||
| 130 | FGF11 | 13918 | -1.484 | 0.0871 | No | ||
| 131 | PDGFB | 13947 | -1.489 | 0.0882 | No | ||
| 132 | MGAT2 | 13981 | -1.500 | 0.0890 | No | ||
| 133 | EPHB1 | 13989 | -1.503 | 0.0911 | No | ||
| 134 | ASCL1 | 14234 | -1.578 | 0.0824 | No | ||
| 135 | PLCB3 | 14401 | -1.621 | 0.0774 | No | ||
| 136 | LRFN5 | 14484 | -1.641 | 0.0763 | No | ||
| 137 | APLP1 | 15082 | -1.827 | 0.0517 | No | ||
| 138 | C1ORF24 | 15251 | -1.882 | 0.0470 | No | ||
| 139 | RAP1GDS1 | 15311 | -1.898 | 0.0473 | No | ||
| 140 | TFEB | 15407 | -1.927 | 0.0461 | No | ||
| 141 | PIP5K2B | 15485 | -1.955 | 0.0457 | No | ||
| 142 | PPP2R5A | 15748 | -2.046 | 0.0369 | No | ||
| 143 | NR2E1 | 15820 | -2.069 | 0.0369 | No | ||
| 144 | DBN1 | 15834 | -2.073 | 0.0396 | No | ||
| 145 | ANGPTL2 | 15841 | -2.076 | 0.0427 | No | ||
| 146 | COL12A1 | 15878 | -2.087 | 0.0444 | No | ||
| 147 | ACVR2B | 16289 | -2.215 | 0.0291 | No | ||
| 148 | FKBP14 | 16480 | -2.284 | 0.0240 | No | ||
| 149 | KCNQ4 | 16519 | -2.295 | 0.0259 | No | ||
| 150 | MYH10 | 16564 | -2.311 | 0.0276 | No | ||
| 151 | CPEB3 | 16628 | -2.332 | 0.0284 | No | ||
| 152 | CENTG2 | 16724 | -2.367 | 0.0278 | No | ||
| 153 | HPN | 16886 | -2.426 | 0.0243 | No | ||
| 154 | EIF4G2 | 16984 | -2.461 | 0.0238 | No | ||
| 155 | BTBD14B | 17149 | -2.516 | 0.0203 | No | ||
| 156 | WNT5A | 17341 | -2.593 | 0.0156 | No | ||
| 157 | KCNAB3 | 17779 | -2.763 | -0.0000 | No | ||
| 158 | OSR2 | 17997 | -2.853 | -0.0054 | No | ||
| 159 | PHF15 | 18005 | -2.856 | -0.0012 | No | ||
| 160 | ST7 | 18077 | -2.889 | 0.0002 | No | ||
| 161 | PTMA | 18360 | -3.006 | -0.0080 | No | ||
| 162 | KCNK3 | 18401 | -3.023 | -0.0050 | No | ||
| 163 | PTK7 | 18730 | -3.202 | -0.0150 | No | ||
| 164 | NCOA6IP | 18754 | -3.213 | -0.0109 | No | ||
| 165 | SMARCD1 | 18797 | -3.235 | -0.0076 | No | ||
| 166 | CCNG1 | 19109 | -3.423 | -0.0165 | No | ||
| 167 | PFKFB3 | 19165 | -3.460 | -0.0134 | No | ||
| 168 | CXXC5 | 19406 | -3.615 | -0.0187 | No | ||
| 169 | ZNRF2 | 19718 | -3.868 | -0.0268 | No | ||
| 170 | ERF | 19734 | -3.887 | -0.0213 | No | ||
| 171 | DLGAP4 | 19907 | -4.040 | -0.0227 | No | ||
| 172 | DGCR8 | 20034 | -4.177 | -0.0218 | No | ||
| 173 | EYA1 | 20145 | -4.286 | -0.0200 | No | ||
| 174 | CGREF1 | 20168 | -4.310 | -0.0142 | No | ||
| 175 | NEO1 | 20318 | -4.470 | -0.0139 | No | ||
| 176 | PABPC1 | 20409 | -4.595 | -0.0107 | No | ||
| 177 | NR2F2 | 20641 | -4.901 | -0.0134 | No | ||
| 178 | HDLBP | 20734 | -5.085 | -0.0095 | No | ||
| 179 | ADIPOR1 | 20887 | -5.303 | -0.0081 | No | ||
| 180 | CIRBP | 20942 | -5.378 | -0.0019 | No | ||
| 181 | SYT7 | 20943 | -5.380 | 0.0067 | No | ||
| 182 | MIB1 | 21173 | -5.895 | 0.0056 | No | ||
| 183 | RGMA | 21205 | -6.003 | 0.0137 | No | ||
| 184 | RPL36AL | 21253 | -6.144 | 0.0214 | No | ||
| 185 | SPATA6 | 21388 | -6.544 | 0.0257 | No |
