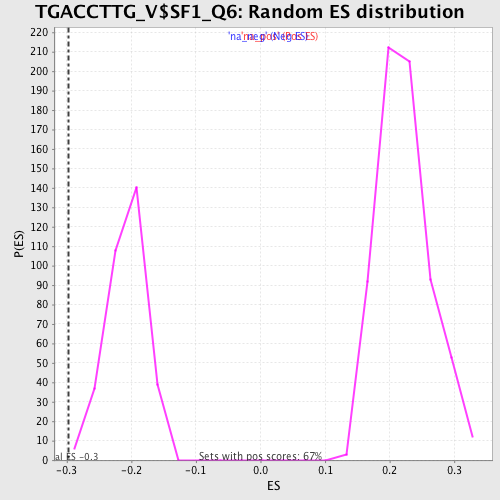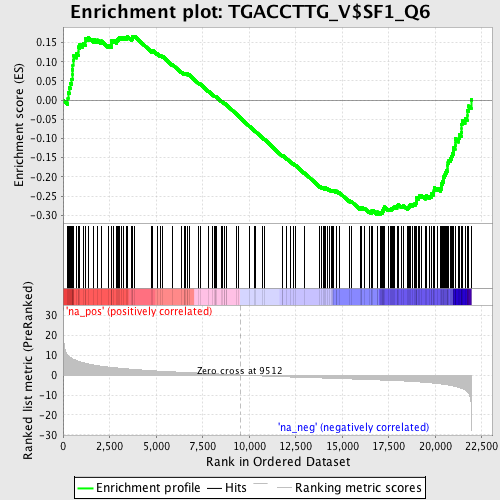
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TGACCTTG_V$SF1_Q6 |
| Enrichment Score (ES) | -0.29793403 |
| Normalized Enrichment Score (NES) | -1.4443725 |
| Nominal p-value | 0.006060606 |
| FDR q-value | 0.18312371 |
| FWER p-Value | 0.957 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLC37A2 | 260 | 9.832 | 0.0042 | No | ||
| 2 | RBMX | 267 | 9.771 | 0.0199 | No | ||
| 3 | MEF2D | 333 | 9.231 | 0.0321 | No | ||
| 4 | ABTB2 | 398 | 8.797 | 0.0436 | No | ||
| 5 | MTCH2 | 455 | 8.423 | 0.0548 | No | ||
| 6 | ARF3 | 491 | 8.188 | 0.0666 | No | ||
| 7 | PPP1R16B | 497 | 8.161 | 0.0798 | No | ||
| 8 | CTSD | 527 | 8.030 | 0.0916 | No | ||
| 9 | ATP6V0C | 532 | 8.011 | 0.1045 | No | ||
| 10 | MSI2 | 575 | 7.805 | 0.1154 | No | ||
| 11 | MADD | 698 | 7.257 | 0.1217 | No | ||
| 12 | GABARAPL1 | 816 | 6.838 | 0.1275 | No | ||
| 13 | SEMA7A | 817 | 6.836 | 0.1387 | No | ||
| 14 | ZNF289 | 899 | 6.605 | 0.1459 | No | ||
| 15 | CKB | 1068 | 6.157 | 0.1482 | No | ||
| 16 | VASP | 1190 | 5.873 | 0.1523 | No | ||
| 17 | UBE1 | 1195 | 5.864 | 0.1617 | No | ||
| 18 | MGST3 | 1353 | 5.500 | 0.1635 | No | ||
| 19 | IBRDC3 | 1629 | 5.005 | 0.1591 | No | ||
| 20 | CRSP7 | 1847 | 4.683 | 0.1568 | No | ||
| 21 | GBAS | 2059 | 4.394 | 0.1543 | No | ||
| 22 | PCTK1 | 2434 | 3.967 | 0.1437 | No | ||
| 23 | CHD2 | 2595 | 3.816 | 0.1426 | No | ||
| 24 | VAMP2 | 2601 | 3.807 | 0.1486 | No | ||
| 25 | CA2 | 2607 | 3.803 | 0.1546 | No | ||
| 26 | PANK1 | 2708 | 3.699 | 0.1561 | No | ||
| 27 | ATP1B1 | 2894 | 3.522 | 0.1533 | No | ||
| 28 | RBMS1 | 2906 | 3.511 | 0.1586 | No | ||
| 29 | HOXC13 | 2966 | 3.464 | 0.1616 | No | ||
| 30 | ESRRA | 3039 | 3.404 | 0.1638 | No | ||
| 31 | DEXI | 3152 | 3.303 | 0.1641 | No | ||
| 32 | RNF139 | 3269 | 3.211 | 0.1640 | No | ||
| 33 | TBX6 | 3402 | 3.103 | 0.1631 | No | ||
| 34 | RANBP10 | 3468 | 3.043 | 0.1651 | No | ||
| 35 | ADCY1 | 3687 | 2.884 | 0.1598 | No | ||
| 36 | COX4I1 | 3729 | 2.857 | 0.1626 | No | ||
| 37 | ATP5B | 3738 | 2.848 | 0.1669 | No | ||
| 38 | FBXO18 | 3843 | 2.771 | 0.1667 | No | ||
| 39 | EPN3 | 4767 | 2.161 | 0.1278 | No | ||
| 40 | CHAT | 4793 | 2.150 | 0.1302 | No | ||
| 41 | POLR3D | 5065 | 1.992 | 0.1210 | No | ||
| 42 | SLC6A8 | 5207 | 1.926 | 0.1177 | No | ||
| 43 | BLCAP | 5363 | 1.850 | 0.1136 | No | ||
| 44 | EPS8L2 | 5886 | 1.598 | 0.0922 | No | ||
| 45 | YWHAH | 6354 | 1.382 | 0.0730 | No | ||
| 46 | MNT | 6512 | 1.303 | 0.0680 | No | ||
| 47 | NCDN | 6518 | 1.301 | 0.0699 | No | ||
| 48 | PDHX | 6575 | 1.272 | 0.0694 | No | ||
| 49 | CASQ2 | 6661 | 1.228 | 0.0675 | No | ||
| 50 | HOXC10 | 6671 | 1.224 | 0.0691 | No | ||
| 51 | RHCG | 6773 | 1.185 | 0.0664 | No | ||
| 52 | GABPA | 7288 | 0.955 | 0.0444 | No | ||
| 53 | KCNG4 | 7367 | 0.924 | 0.0423 | No | ||
| 54 | RAB3A | 7806 | 0.737 | 0.0234 | No | ||
| 55 | UQCRC1 | 8034 | 0.647 | 0.0140 | No | ||
| 56 | ZRANB1 | 8131 | 0.604 | 0.0106 | No | ||
| 57 | RGS7 | 8181 | 0.585 | 0.0093 | No | ||
| 58 | SNTG1 | 8189 | 0.582 | 0.0099 | No | ||
| 59 | TUBB4 | 8221 | 0.568 | 0.0094 | No | ||
| 60 | NXPH4 | 8515 | 0.446 | -0.0033 | No | ||
| 61 | GABRA3 | 8584 | 0.416 | -0.0057 | No | ||
| 62 | AMY2A | 8660 | 0.385 | -0.0085 | No | ||
| 63 | HCN1 | 8694 | 0.372 | -0.0094 | No | ||
| 64 | KIF5A | 8767 | 0.333 | -0.0122 | No | ||
| 65 | MAL | 8784 | 0.327 | -0.0124 | No | ||
| 66 | STAC2 | 9317 | 0.099 | -0.0367 | No | ||
| 67 | ATP6V1B1 | 9422 | 0.047 | -0.0414 | No | ||
| 68 | UCHL1 | 9999 | -0.220 | -0.0675 | No | ||
| 69 | FZD9 | 10258 | -0.319 | -0.0788 | No | ||
| 70 | PNOC | 10333 | -0.352 | -0.0816 | No | ||
| 71 | ATP5G3 | 10692 | -0.472 | -0.0973 | No | ||
| 72 | SLC30A3 | 10803 | -0.513 | -0.1015 | No | ||
| 73 | CHGA | 11789 | -0.853 | -0.1453 | No | ||
| 74 | CDH16 | 11811 | -0.860 | -0.1449 | No | ||
| 75 | UBL3 | 12011 | -0.924 | -0.1525 | No | ||
| 76 | KCNMB2 | 12236 | -0.987 | -0.1612 | No | ||
| 77 | FNDC5 | 12379 | -1.031 | -0.1660 | No | ||
| 78 | ZDHHC21 | 12495 | -1.067 | -0.1696 | No | ||
| 79 | FGF9 | 12977 | -1.209 | -0.1897 | No | ||
| 80 | SCN5A | 13788 | -1.447 | -0.2245 | No | ||
| 81 | CACNB2 | 13894 | -1.477 | -0.2269 | No | ||
| 82 | ANKRD28 | 13973 | -1.498 | -0.2280 | No | ||
| 83 | NTF3 | 14066 | -1.527 | -0.2297 | No | ||
| 84 | INSR | 14072 | -1.529 | -0.2275 | No | ||
| 85 | MAEA | 14193 | -1.566 | -0.2304 | No | ||
| 86 | SLC38A3 | 14310 | -1.600 | -0.2331 | No | ||
| 87 | PHACTR3 | 14403 | -1.621 | -0.2347 | No | ||
| 88 | LRFN5 | 14484 | -1.641 | -0.2357 | No | ||
| 89 | ELAVL3 | 14529 | -1.656 | -0.2350 | No | ||
| 90 | RILP | 14676 | -1.703 | -0.2389 | No | ||
| 91 | SV2A | 14689 | -1.708 | -0.2366 | No | ||
| 92 | TCF2 | 14838 | -1.752 | -0.2406 | No | ||
| 93 | FANCD2 | 15380 | -1.921 | -0.2623 | No | ||
| 94 | DSCR1L1 | 15475 | -1.950 | -0.2634 | No | ||
| 95 | HSPE1 | 15969 | -2.115 | -0.2826 | No | ||
| 96 | CNTN2 | 16016 | -2.126 | -0.2812 | No | ||
| 97 | MYL3 | 16041 | -2.134 | -0.2788 | No | ||
| 98 | SH3GL2 | 16184 | -2.175 | -0.2818 | No | ||
| 99 | ABHD3 | 16450 | -2.270 | -0.2902 | No | ||
| 100 | CCNE1 | 16580 | -2.317 | -0.2923 | No | ||
| 101 | TBC1D15 | 16586 | -2.320 | -0.2888 | No | ||
| 102 | SLITRK1 | 16616 | -2.328 | -0.2863 | No | ||
| 103 | LAMB2 | 16871 | -2.420 | -0.2940 | Yes | ||
| 104 | ATP5C1 | 16900 | -2.431 | -0.2913 | Yes | ||
| 105 | NNAT | 17039 | -2.479 | -0.2935 | Yes | ||
| 106 | PCDH7 | 17094 | -2.497 | -0.2919 | Yes | ||
| 107 | EEF1A2 | 17157 | -2.518 | -0.2906 | Yes | ||
| 108 | KIN | 17169 | -2.522 | -0.2870 | Yes | ||
| 109 | MEF2C | 17220 | -2.542 | -0.2851 | Yes | ||
| 110 | CX3CL1 | 17229 | -2.546 | -0.2813 | Yes | ||
| 111 | RPH3A | 17250 | -2.554 | -0.2781 | Yes | ||
| 112 | RAPGEF5 | 17471 | -2.638 | -0.2839 | Yes | ||
| 113 | SLC2A4 | 17567 | -2.667 | -0.2838 | Yes | ||
| 114 | NRXN2 | 17628 | -2.692 | -0.2822 | Yes | ||
| 115 | ASTN2 | 17710 | -2.731 | -0.2814 | Yes | ||
| 116 | VSNL1 | 17748 | -2.748 | -0.2786 | Yes | ||
| 117 | CYC1 | 17827 | -2.784 | -0.2776 | Yes | ||
| 118 | ATP6AP2 | 17941 | -2.830 | -0.2782 | Yes | ||
| 119 | KCNK1 | 17947 | -2.832 | -0.2738 | Yes | ||
| 120 | PHF15 | 18005 | -2.856 | -0.2717 | Yes | ||
| 121 | RHOT1 | 18204 | -2.937 | -0.2760 | Yes | ||
| 122 | NTNG2 | 18288 | -2.970 | -0.2749 | Yes | ||
| 123 | STK32A | 18526 | -3.086 | -0.2808 | Yes | ||
| 124 | SEMA3F | 18584 | -3.113 | -0.2783 | Yes | ||
| 125 | CATSPER2 | 18597 | -3.123 | -0.2737 | Yes | ||
| 126 | CEL | 18648 | -3.149 | -0.2708 | Yes | ||
| 127 | DNAJC11 | 18766 | -3.221 | -0.2709 | Yes | ||
| 128 | HTF9C | 18872 | -3.275 | -0.2704 | Yes | ||
| 129 | HMGB3 | 18940 | -3.321 | -0.2680 | Yes | ||
| 130 | MASP1 | 18974 | -3.339 | -0.2641 | Yes | ||
| 131 | AUH | 18989 | -3.348 | -0.2592 | Yes | ||
| 132 | TMEM28 | 18991 | -3.349 | -0.2538 | Yes | ||
| 133 | SDK1 | 19103 | -3.419 | -0.2533 | Yes | ||
| 134 | DNER | 19141 | -3.446 | -0.2493 | Yes | ||
| 135 | COX5B | 19264 | -3.515 | -0.2492 | Yes | ||
| 136 | KIRREL3 | 19452 | -3.649 | -0.2518 | Yes | ||
| 137 | DIRAS1 | 19507 | -3.694 | -0.2482 | Yes | ||
| 138 | NDUFB5 | 19693 | -3.849 | -0.2504 | Yes | ||
| 139 | MTX2 | 19794 | -3.941 | -0.2485 | Yes | ||
| 140 | WDFY3 | 19807 | -3.956 | -0.2426 | Yes | ||
| 141 | ACO2 | 19914 | -4.047 | -0.2408 | Yes | ||
| 142 | ATP5J | 19927 | -4.060 | -0.2347 | Yes | ||
| 143 | WWP1 | 19929 | -4.061 | -0.2281 | Yes | ||
| 144 | TRAP1 | 20113 | -4.258 | -0.2295 | Yes | ||
| 145 | TNNI1 | 20281 | -4.430 | -0.2299 | Yes | ||
| 146 | SNPH | 20327 | -4.483 | -0.2246 | Yes | ||
| 147 | MRPL47 | 20335 | -4.490 | -0.2176 | Yes | ||
| 148 | NPTX2 | 20379 | -4.547 | -0.2121 | Yes | ||
| 149 | SYNGR1 | 20429 | -4.632 | -0.2068 | Yes | ||
| 150 | RNF4 | 20447 | -4.652 | -0.1999 | Yes | ||
| 151 | PRPSAP1 | 20508 | -4.731 | -0.1949 | Yes | ||
| 152 | AFG3L2 | 20555 | -4.780 | -0.1892 | Yes | ||
| 153 | AK2 | 20581 | -4.814 | -0.1825 | Yes | ||
| 154 | APP | 20670 | -4.957 | -0.1784 | Yes | ||
| 155 | CS | 20672 | -4.961 | -0.1703 | Yes | ||
| 156 | COMTD1 | 20679 | -4.971 | -0.1624 | Yes | ||
| 157 | PHF5A | 20728 | -5.077 | -0.1563 | Yes | ||
| 158 | FSTL3 | 20830 | -5.234 | -0.1524 | Yes | ||
| 159 | RIN2 | 20876 | -5.291 | -0.1457 | Yes | ||
| 160 | HSPD1 | 20908 | -5.331 | -0.1384 | Yes | ||
| 161 | TFRC | 20958 | -5.404 | -0.1318 | Yes | ||
| 162 | RREB1 | 20991 | -5.466 | -0.1243 | Yes | ||
| 163 | VDAC2 | 21060 | -5.633 | -0.1182 | Yes | ||
| 164 | RANBP1 | 21068 | -5.642 | -0.1093 | Yes | ||
| 165 | ALDOA | 21071 | -5.645 | -0.1001 | Yes | ||
| 166 | CAMK2G | 21269 | -6.196 | -0.0990 | Yes | ||
| 167 | NDFIP1 | 21290 | -6.264 | -0.0897 | Yes | ||
| 168 | SDHD | 21390 | -6.548 | -0.0835 | Yes | ||
| 169 | ATP5G1 | 21410 | -6.613 | -0.0735 | Yes | ||
| 170 | TIMM8B | 21413 | -6.619 | -0.0627 | Yes | ||
| 171 | PPP3CC | 21439 | -6.721 | -0.0529 | Yes | ||
| 172 | IMMT | 21596 | -7.320 | -0.0480 | Yes | ||
| 173 | TOMM40 | 21709 | -8.157 | -0.0398 | Yes | ||
| 174 | TIMM9 | 21713 | -8.175 | -0.0265 | Yes | ||
| 175 | IDH3A | 21783 | -8.933 | -0.0151 | Yes | ||
| 176 | AMPD3 | 21923 | -13.760 | 0.0011 | Yes |