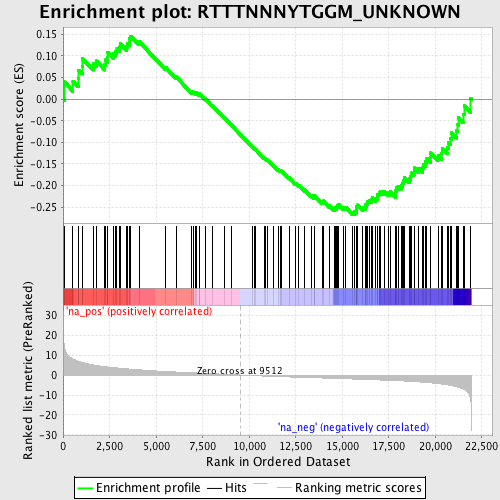
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | RTTTNNNYTGGM_UNKNOWN |
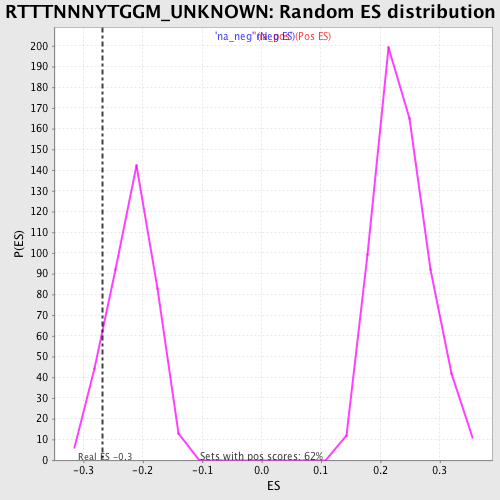
| Enrichment Score (ES) | -0.26718 |
| Normalized Enrichment Score (NES) | -1.2231281 |
| Nominal p-value | 0.10526316 |
| FDR q-value | 0.4086131 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DHRS3 | 61 | 14.736 | 0.0399 | No | ||
| 2 | STX6 | 529 | 8.020 | 0.0418 | No | ||
| 3 | NFATC1 | 826 | 6.800 | 0.0479 | No | ||
| 4 | BACH1 | 833 | 6.787 | 0.0673 | No | ||
| 5 | ODF2 | 1032 | 6.258 | 0.0764 | No | ||
| 6 | FOXP1 | 1045 | 6.223 | 0.0939 | No | ||
| 7 | USP7 | 1623 | 5.012 | 0.0820 | No | ||
| 8 | DSCR1 | 1784 | 4.781 | 0.0885 | No | ||
| 9 | INPP5A | 2244 | 4.176 | 0.0796 | No | ||
| 10 | CFL2 | 2263 | 4.155 | 0.0908 | No | ||
| 11 | MAP2K6 | 2378 | 4.033 | 0.0972 | No | ||
| 12 | OGT | 2385 | 4.030 | 0.1086 | No | ||
| 13 | DCX | 2692 | 3.710 | 0.1054 | No | ||
| 14 | PPP2R5E | 2818 | 3.598 | 0.1101 | No | ||
| 15 | ATP1B1 | 2894 | 3.522 | 0.1169 | No | ||
| 16 | TRPS1 | 3024 | 3.417 | 0.1208 | No | ||
| 17 | BCL11A | 3071 | 3.369 | 0.1285 | No | ||
| 18 | TANK | 3424 | 3.086 | 0.1213 | No | ||
| 19 | AP1G2 | 3462 | 3.050 | 0.1285 | No | ||
| 20 | RFX3 | 3564 | 2.971 | 0.1324 | No | ||
| 21 | KLF5 | 3581 | 2.962 | 0.1403 | No | ||
| 22 | UNC50 | 3642 | 2.912 | 0.1460 | No | ||
| 23 | FBXO11 | 4085 | 2.599 | 0.1333 | No | ||
| 24 | PTPRCAP | 5519 | 1.769 | 0.0728 | No | ||
| 25 | PUM2 | 6074 | 1.509 | 0.0518 | No | ||
| 26 | F13A1 | 6912 | 1.117 | 0.0167 | No | ||
| 27 | SPAG9 | 6982 | 1.088 | 0.0166 | No | ||
| 28 | MITF | 7130 | 1.025 | 0.0129 | No | ||
| 29 | REPS2 | 7150 | 1.017 | 0.0150 | No | ||
| 30 | PALM2 | 7315 | 0.944 | 0.0102 | No | ||
| 31 | IL23A | 7322 | 0.940 | 0.0126 | No | ||
| 32 | MIP | 7637 | 0.813 | 0.0006 | No | ||
| 33 | RBP4 | 8026 | 0.650 | -0.0153 | No | ||
| 34 | ACCN1 | 8657 | 0.386 | -0.0430 | No | ||
| 35 | TGM5 | 9073 | 0.200 | -0.0614 | No | ||
| 36 | PPP1R1B | 10155 | -0.280 | -0.1102 | No | ||
| 37 | HOXA11 | 10259 | -0.319 | -0.1139 | No | ||
| 38 | SERPIND1 | 10342 | -0.355 | -0.1167 | No | ||
| 39 | ATP10D | 10804 | -0.514 | -0.1363 | No | ||
| 40 | SLC6A9 | 10859 | -0.532 | -0.1372 | No | ||
| 41 | AQP5 | 10976 | -0.574 | -0.1409 | No | ||
| 42 | HOXC4 | 11324 | -0.689 | -0.1548 | No | ||
| 43 | TRIM47 | 11559 | -0.771 | -0.1633 | No | ||
| 44 | DMXL1 | 11662 | -0.807 | -0.1656 | No | ||
| 45 | ATPAF2 | 11731 | -0.837 | -0.1663 | No | ||
| 46 | ZIC2 | 12142 | -0.959 | -0.1823 | No | ||
| 47 | PCYT1B | 12474 | -1.057 | -0.1944 | No | ||
| 48 | PDGFRL | 12669 | -1.114 | -0.2000 | No | ||
| 49 | PITX1 | 12964 | -1.204 | -0.2100 | No | ||
| 50 | ZIC1 | 13330 | -1.309 | -0.2229 | No | ||
| 51 | TCF1 | 13483 | -1.356 | -0.2260 | No | ||
| 52 | FHIT | 13488 | -1.358 | -0.2222 | No | ||
| 53 | NOS3 | 13915 | -1.482 | -0.2374 | No | ||
| 54 | ANKRD28 | 13973 | -1.498 | -0.2357 | No | ||
| 55 | SLC38A3 | 14310 | -1.600 | -0.2465 | No | ||
| 56 | NAV3 | 14566 | -1.670 | -0.2533 | No | ||
| 57 | PAIP2 | 14627 | -1.690 | -0.2512 | No | ||
| 58 | GABRG2 | 14706 | -1.713 | -0.2498 | No | ||
| 59 | FBLN5 | 14756 | -1.729 | -0.2470 | No | ||
| 60 | CALM2 | 14820 | -1.748 | -0.2448 | No | ||
| 61 | GCM1 | 15078 | -1.826 | -0.2513 | No | ||
| 62 | SLCO2A1 | 15172 | -1.858 | -0.2502 | No | ||
| 63 | CAB39L | 15544 | -1.979 | -0.2614 | Yes | ||
| 64 | HOXC6 | 15663 | -2.021 | -0.2610 | Yes | ||
| 65 | ETV1 | 15743 | -2.044 | -0.2587 | Yes | ||
| 66 | HOXA2 | 15771 | -2.054 | -0.2540 | Yes | ||
| 67 | HOXD3 | 15779 | -2.056 | -0.2483 | Yes | ||
| 68 | NGFB | 15842 | -2.076 | -0.2452 | Yes | ||
| 69 | HOXA7 | 16112 | -2.156 | -0.2512 | Yes | ||
| 70 | PROK2 | 16232 | -2.196 | -0.2503 | Yes | ||
| 71 | ESR1 | 16252 | -2.203 | -0.2448 | Yes | ||
| 72 | SLC31A2 | 16327 | -2.224 | -0.2418 | Yes | ||
| 73 | PHEX | 16375 | -2.241 | -0.2374 | Yes | ||
| 74 | HAPLN1 | 16441 | -2.267 | -0.2338 | Yes | ||
| 75 | ALDH1A2 | 16563 | -2.311 | -0.2327 | Yes | ||
| 76 | HAS2 | 16601 | -2.323 | -0.2276 | Yes | ||
| 77 | DMD | 16796 | -2.392 | -0.2296 | Yes | ||
| 78 | HPN | 16886 | -2.426 | -0.2266 | Yes | ||
| 79 | ACACA | 16902 | -2.431 | -0.2203 | Yes | ||
| 80 | TECTA | 16974 | -2.457 | -0.2164 | Yes | ||
| 81 | CALD1 | 17060 | -2.487 | -0.2131 | Yes | ||
| 82 | DPF3 | 17243 | -2.551 | -0.2140 | Yes | ||
| 83 | BOK | 17462 | -2.635 | -0.2164 | Yes | ||
| 84 | UNC13B | 17589 | -2.679 | -0.2144 | Yes | ||
| 85 | ZADH2 | 17870 | -2.796 | -0.2191 | Yes | ||
| 86 | ZIC4 | 17876 | -2.800 | -0.2112 | Yes | ||
| 87 | FOXP2 | 17925 | -2.821 | -0.2052 | Yes | ||
| 88 | CLDN14 | 18035 | -2.870 | -0.2019 | Yes | ||
| 89 | DQX1 | 18177 | -2.928 | -0.1999 | Yes | ||
| 90 | EGFR | 18247 | -2.953 | -0.1945 | Yes | ||
| 91 | BMP5 | 18290 | -2.971 | -0.1878 | Yes | ||
| 92 | FXYD5 | 18344 | -2.999 | -0.1816 | Yes | ||
| 93 | NTRK2 | 18614 | -3.135 | -0.1848 | Yes | ||
| 94 | RGS3 | 18672 | -3.160 | -0.1782 | Yes | ||
| 95 | PTPRG | 18716 | -3.190 | -0.1710 | Yes | ||
| 96 | GRIA1 | 18857 | -3.266 | -0.1679 | Yes | ||
| 97 | CREB5 | 18878 | -3.278 | -0.1593 | Yes | ||
| 98 | RPP21 | 19104 | -3.419 | -0.1597 | Yes | ||
| 99 | PLUNC | 19321 | -3.548 | -0.1593 | Yes | ||
| 100 | GGA2 | 19352 | -3.579 | -0.1503 | Yes | ||
| 101 | DDX46 | 19467 | -3.662 | -0.1449 | Yes | ||
| 102 | NFATC2 | 19548 | -3.725 | -0.1378 | Yes | ||
| 103 | IMPDH2 | 19737 | -3.887 | -0.1352 | Yes | ||
| 104 | GPR85 | 19754 | -3.901 | -0.1246 | Yes | ||
| 105 | EYA1 | 20145 | -4.286 | -0.1300 | Yes | ||
| 106 | MARCO | 20343 | -4.502 | -0.1260 | Yes | ||
| 107 | BRUNOL4 | 20358 | -4.522 | -0.1135 | Yes | ||
| 108 | LRP1 | 20637 | -4.891 | -0.1121 | Yes | ||
| 109 | NFIB | 20706 | -5.034 | -0.1006 | Yes | ||
| 110 | LMO4 | 20804 | -5.199 | -0.0900 | Yes | ||
| 111 | MAB21L2 | 20881 | -5.298 | -0.0781 | Yes | ||
| 112 | AKAP2 | 21153 | -5.839 | -0.0736 | Yes | ||
| 113 | SLC11A2 | 21206 | -6.004 | -0.0586 | Yes | ||
| 114 | SWAP70 | 21250 | -6.137 | -0.0428 | Yes | ||
| 115 | PTP4A1 | 21516 | -7.000 | -0.0346 | Yes | ||
| 116 | KCTD6 | 21550 | -7.119 | -0.0155 | Yes | ||
| 117 | ID2 | 21890 | -11.617 | 0.0026 | Yes |