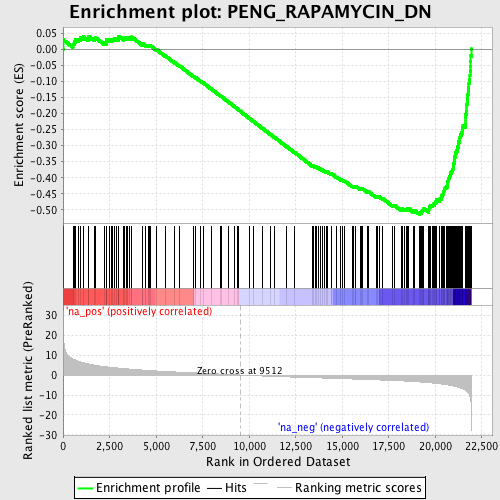
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | PENG_RAPAMYCIN_DN |
| Enrichment Score (ES) | -0.5141355 |
| Normalized Enrichment Score (NES) | -2.5148683 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | XBP1 | 6 | 21.674 | 0.0291 | No | ||
| 2 | ATP6V0C | 532 | 8.011 | 0.0159 | No | ||
| 3 | LIMK2 | 596 | 7.693 | 0.0234 | No | ||
| 4 | LDHB | 657 | 7.455 | 0.0307 | No | ||
| 5 | PAPOLA | 846 | 6.743 | 0.0312 | No | ||
| 6 | INPP5E | 924 | 6.529 | 0.0366 | No | ||
| 7 | SAFB | 1070 | 6.151 | 0.0382 | No | ||
| 8 | SREBF1 | 1355 | 5.494 | 0.0326 | No | ||
| 9 | GTF3C1 | 1384 | 5.424 | 0.0387 | No | ||
| 10 | RAB1A | 1676 | 4.929 | 0.0320 | No | ||
| 11 | XPO6 | 1747 | 4.828 | 0.0354 | No | ||
| 12 | RTN4 | 2205 | 4.221 | 0.0201 | No | ||
| 13 | ICT1 | 2310 | 4.104 | 0.0209 | No | ||
| 14 | PIP5K1A | 2320 | 4.092 | 0.0260 | No | ||
| 15 | BCAP31 | 2330 | 4.084 | 0.0311 | No | ||
| 16 | PICALM | 2482 | 3.908 | 0.0295 | No | ||
| 17 | DHCR24 | 2610 | 3.802 | 0.0288 | No | ||
| 18 | GSPT1 | 2674 | 3.729 | 0.0310 | No | ||
| 19 | EXTL3 | 2758 | 3.653 | 0.0321 | No | ||
| 20 | SLC1A5 | 2865 | 3.549 | 0.0321 | No | ||
| 21 | CD47 | 2977 | 3.454 | 0.0316 | No | ||
| 22 | WDR18 | 2984 | 3.449 | 0.0360 | No | ||
| 23 | HADH2 | 2993 | 3.443 | 0.0403 | No | ||
| 24 | BMP1 | 3260 | 3.219 | 0.0325 | No | ||
| 25 | UBE2M | 3308 | 3.171 | 0.0346 | No | ||
| 26 | ALDH4A1 | 3396 | 3.105 | 0.0348 | No | ||
| 27 | PTBP1 | 3472 | 3.039 | 0.0355 | No | ||
| 28 | ATP6V0E | 3568 | 2.969 | 0.0352 | No | ||
| 29 | ARF1 | 3666 | 2.899 | 0.0347 | No | ||
| 30 | RAB14 | 3685 | 2.885 | 0.0377 | No | ||
| 31 | FKBP4 | 4279 | 2.454 | 0.0138 | No | ||
| 32 | MMP15 | 4281 | 2.453 | 0.0171 | No | ||
| 33 | PTPN1 | 4451 | 2.352 | 0.0125 | No | ||
| 34 | MAPKAPK3 | 4580 | 2.275 | 0.0097 | No | ||
| 35 | CHERP | 4624 | 2.242 | 0.0108 | No | ||
| 36 | RBBP6 | 4707 | 2.195 | 0.0100 | No | ||
| 37 | COX11 | 5011 | 2.027 | -0.0012 | No | ||
| 38 | HYOU1 | 5474 | 1.790 | -0.0199 | No | ||
| 39 | NDUFA9 | 5990 | 1.549 | -0.0415 | No | ||
| 40 | MLF2 | 6279 | 1.415 | -0.0528 | No | ||
| 41 | CHUK | 6991 | 1.084 | -0.0840 | No | ||
| 42 | TUBG1 | 7124 | 1.028 | -0.0887 | No | ||
| 43 | GCDH | 7381 | 0.917 | -0.0992 | No | ||
| 44 | EIF2B2 | 7525 | 0.859 | -0.1046 | No | ||
| 45 | TGDS | 7963 | 0.676 | -0.1237 | No | ||
| 46 | PSMB5 | 8431 | 0.485 | -0.1445 | No | ||
| 47 | DAP3 | 8494 | 0.455 | -0.1467 | No | ||
| 48 | USP10 | 8859 | 0.296 | -0.1631 | No | ||
| 49 | EMP3 | 9212 | 0.142 | -0.1790 | No | ||
| 50 | EIF2B4 | 9387 | 0.064 | -0.1869 | No | ||
| 51 | BCS1L | 9410 | 0.051 | -0.1879 | No | ||
| 52 | UCHL1 | 9999 | -0.220 | -0.2146 | No | ||
| 53 | ACAT2 | 10203 | -0.297 | -0.2235 | No | ||
| 54 | ATP5G3 | 10692 | -0.472 | -0.2453 | No | ||
| 55 | PPP2R2A | 11150 | -0.638 | -0.2654 | No | ||
| 56 | CAPZA2 | 11367 | -0.704 | -0.2744 | No | ||
| 57 | BTK | 12025 | -0.929 | -0.3033 | No | ||
| 58 | EBP | 12446 | -1.049 | -0.3211 | No | ||
| 59 | NP | 13376 | -1.324 | -0.3620 | No | ||
| 60 | WNT10B | 13428 | -1.341 | -0.3625 | No | ||
| 61 | RPS26 | 13546 | -1.378 | -0.3660 | No | ||
| 62 | JTB | 13589 | -1.389 | -0.3661 | No | ||
| 63 | SDHB | 13698 | -1.428 | -0.3691 | No | ||
| 64 | AHSA1 | 13811 | -1.454 | -0.3723 | No | ||
| 65 | CSE1L | 13929 | -1.485 | -0.3757 | No | ||
| 66 | EIF4G1 | 14061 | -1.526 | -0.3796 | No | ||
| 67 | PPIB | 14131 | -1.543 | -0.3807 | No | ||
| 68 | PSMD8 | 14190 | -1.564 | -0.3812 | No | ||
| 69 | S100A4 | 14405 | -1.622 | -0.3888 | No | ||
| 70 | DDX3X | 14411 | -1.623 | -0.3869 | No | ||
| 71 | MRPS12 | 14711 | -1.715 | -0.3983 | No | ||
| 72 | FASN | 14913 | -1.774 | -0.4051 | No | ||
| 73 | CALU | 15009 | -1.803 | -0.4070 | No | ||
| 74 | BYSL | 15131 | -1.846 | -0.4101 | No | ||
| 75 | HCCS | 15536 | -1.975 | -0.4260 | No | ||
| 76 | WBSCR22 | 15604 | -2.001 | -0.4263 | No | ||
| 77 | PFKM | 15687 | -2.030 | -0.4273 | No | ||
| 78 | POLR2B | 15722 | -2.038 | -0.4261 | No | ||
| 79 | HSPE1 | 15969 | -2.115 | -0.4346 | No | ||
| 80 | YARS | 16024 | -2.129 | -0.4342 | No | ||
| 81 | PTRF | 16074 | -2.145 | -0.4335 | No | ||
| 82 | RNASEH1 | 16361 | -2.237 | -0.4436 | No | ||
| 83 | IFI30 | 16382 | -2.243 | -0.4415 | No | ||
| 84 | COPB2 | 16829 | -2.406 | -0.4587 | No | ||
| 85 | NXF1 | 16911 | -2.434 | -0.4591 | No | ||
| 86 | EIF4G2 | 16984 | -2.461 | -0.4591 | No | ||
| 87 | HSPA5 | 17144 | -2.514 | -0.4630 | No | ||
| 88 | IDI1 | 17723 | -2.738 | -0.4858 | No | ||
| 89 | CYC1 | 17827 | -2.784 | -0.4868 | No | ||
| 90 | SLC39A14 | 18183 | -2.929 | -0.4991 | No | ||
| 91 | P2RX5 | 18213 | -2.940 | -0.4965 | No | ||
| 92 | RABGGTB | 18362 | -3.006 | -0.4992 | No | ||
| 93 | NDUFS3 | 18441 | -3.046 | -0.4986 | No | ||
| 94 | POLR2F | 18484 | -3.064 | -0.4964 | No | ||
| 95 | SLC3A2 | 18581 | -3.111 | -0.4966 | No | ||
| 96 | SSBP1 | 18821 | -3.245 | -0.5032 | No | ||
| 97 | SYNCRIP | 18869 | -3.273 | -0.5009 | No | ||
| 98 | RNU3IP2 | 19158 | -3.456 | -0.5095 | Yes | ||
| 99 | GM2A | 19210 | -3.485 | -0.5071 | Yes | ||
| 100 | MARS | 19273 | -3.521 | -0.5051 | Yes | ||
| 101 | ATP1B3 | 19291 | -3.533 | -0.5011 | Yes | ||
| 102 | ACVRL1 | 19338 | -3.567 | -0.4984 | Yes | ||
| 103 | NUP98 | 19385 | -3.602 | -0.4956 | Yes | ||
| 104 | CCNC | 19654 | -3.809 | -0.5028 | Yes | ||
| 105 | NOL1 | 19659 | -3.816 | -0.4978 | Yes | ||
| 106 | SCD | 19662 | -3.819 | -0.4927 | Yes | ||
| 107 | MORF4L2 | 19680 | -3.832 | -0.4883 | Yes | ||
| 108 | ARMET | 19739 | -3.891 | -0.4857 | Yes | ||
| 109 | ASNS | 19842 | -3.983 | -0.4850 | Yes | ||
| 110 | GCN5L2 | 19885 | -4.021 | -0.4815 | Yes | ||
| 111 | MYCBP | 19976 | -4.104 | -0.4800 | Yes | ||
| 112 | SQLE | 19992 | -4.126 | -0.4751 | Yes | ||
| 113 | PTDSS1 | 20056 | -4.198 | -0.4723 | Yes | ||
| 114 | CCT2 | 20075 | -4.210 | -0.4674 | Yes | ||
| 115 | DNAJA1 | 20208 | -4.367 | -0.4676 | Yes | ||
| 116 | IDH3B | 20250 | -4.402 | -0.4635 | Yes | ||
| 117 | CCT6A | 20314 | -4.466 | -0.4603 | Yes | ||
| 118 | GTF2A2 | 20323 | -4.477 | -0.4546 | Yes | ||
| 119 | NMT1 | 20406 | -4.591 | -0.4522 | Yes | ||
| 120 | SF3A3 | 20420 | -4.616 | -0.4465 | Yes | ||
| 121 | EEF1E1 | 20455 | -4.662 | -0.4418 | Yes | ||
| 122 | RAE1 | 20469 | -4.687 | -0.4360 | Yes | ||
| 123 | GLO1 | 20518 | -4.742 | -0.4318 | Yes | ||
| 124 | AK2 | 20581 | -4.814 | -0.4281 | Yes | ||
| 125 | CYCS | 20634 | -4.887 | -0.4239 | Yes | ||
| 126 | KPNA3 | 20663 | -4.935 | -0.4185 | Yes | ||
| 127 | ADRM1 | 20665 | -4.939 | -0.4118 | Yes | ||
| 128 | CDC25A | 20686 | -4.987 | -0.4060 | Yes | ||
| 129 | PRDX3 | 20726 | -5.075 | -0.4009 | Yes | ||
| 130 | MRPL12 | 20741 | -5.100 | -0.3946 | Yes | ||
| 131 | ETFA | 20794 | -5.187 | -0.3900 | Yes | ||
| 132 | CCT5 | 20836 | -5.245 | -0.3847 | Yes | ||
| 133 | EIF3S9 | 20874 | -5.289 | -0.3793 | Yes | ||
| 134 | PSMD5 | 20934 | -5.370 | -0.3747 | Yes | ||
| 135 | TRIP13 | 20952 | -5.396 | -0.3682 | Yes | ||
| 136 | TFRC | 20958 | -5.404 | -0.3611 | Yes | ||
| 137 | CCT4 | 20972 | -5.429 | -0.3543 | Yes | ||
| 138 | TARS | 21015 | -5.523 | -0.3488 | Yes | ||
| 139 | CDK4 | 21020 | -5.528 | -0.3414 | Yes | ||
| 140 | ETF1 | 21034 | -5.559 | -0.3345 | Yes | ||
| 141 | HSPA4 | 21067 | -5.641 | -0.3283 | Yes | ||
| 142 | SRM | 21080 | -5.666 | -0.3212 | Yes | ||
| 143 | UBE2L3 | 21148 | -5.829 | -0.3164 | Yes | ||
| 144 | PSMD14 | 21175 | -5.898 | -0.3096 | Yes | ||
| 145 | PITPNB | 21208 | -6.006 | -0.3029 | Yes | ||
| 146 | PPIF | 21245 | -6.125 | -0.2963 | Yes | ||
| 147 | TCP1 | 21257 | -6.160 | -0.2884 | Yes | ||
| 148 | RTCD1 | 21314 | -6.331 | -0.2824 | Yes | ||
| 149 | PSMD11 | 21321 | -6.341 | -0.2741 | Yes | ||
| 150 | NME1 | 21334 | -6.394 | -0.2660 | Yes | ||
| 151 | TIMM17A | 21399 | -6.579 | -0.2600 | Yes | ||
| 152 | NARS | 21438 | -6.717 | -0.2526 | Yes | ||
| 153 | PHB | 21449 | -6.759 | -0.2439 | Yes | ||
| 154 | CTSC | 21486 | -6.873 | -0.2363 | Yes | ||
| 155 | PSMC2 | 21627 | -7.542 | -0.2325 | Yes | ||
| 156 | AMD1 | 21631 | -7.566 | -0.2223 | Yes | ||
| 157 | SLC29A1 | 21634 | -7.574 | -0.2122 | Yes | ||
| 158 | ATF5 | 21640 | -7.604 | -0.2021 | Yes | ||
| 159 | CTPS | 21657 | -7.720 | -0.1924 | Yes | ||
| 160 | TIMM44 | 21690 | -7.999 | -0.1830 | Yes | ||
| 161 | RARS | 21692 | -8.005 | -0.1722 | Yes | ||
| 162 | TOMM40 | 21709 | -8.157 | -0.1619 | Yes | ||
| 163 | PA2G4 | 21722 | -8.235 | -0.1512 | Yes | ||
| 164 | EBNA1BP2 | 21747 | -8.506 | -0.1408 | Yes | ||
| 165 | GORASP2 | 21765 | -8.721 | -0.1298 | Yes | ||
| 166 | IDH3A | 21783 | -8.933 | -0.1184 | Yes | ||
| 167 | AATF | 21801 | -9.220 | -0.1067 | Yes | ||
| 168 | DDX21 | 21826 | -9.585 | -0.0948 | Yes | ||
| 169 | NOL5A | 21859 | -10.573 | -0.0820 | Yes | ||
| 170 | SNRPA1 | 21866 | -10.806 | -0.0676 | Yes | ||
| 171 | SLC7A5 | 21884 | -11.489 | -0.0528 | Yes | ||
| 172 | ID2 | 21890 | -11.617 | -0.0373 | Yes | ||
| 173 | GOT2 | 21912 | -12.841 | -0.0209 | Yes | ||
| 174 | C1QBP | 21938 | -16.533 | 0.0004 | Yes |
