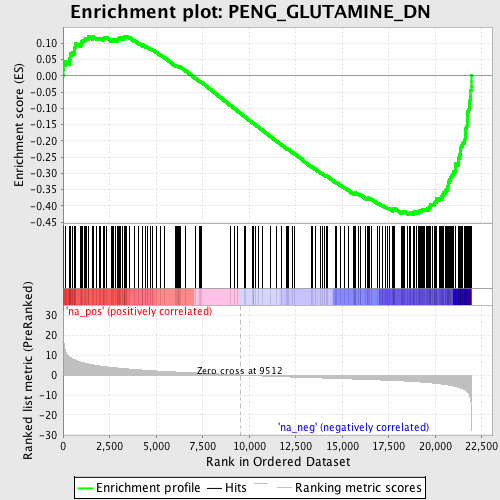
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | PENG_GLUTAMINE_DN |
| Enrichment Score (ES) | -0.42641976 |
| Normalized Enrichment Score (NES) | -2.1721985 |
| Nominal p-value | 0.0 |
| FDR q-value | 8.8633137E-4 |
| FWER p-Value | 0.017 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | XBP1 | 6 | 21.674 | 0.0210 | No | ||
| 2 | HS6ST1 | 35 | 17.027 | 0.0364 | No | ||
| 3 | DNAJB1 | 114 | 12.542 | 0.0451 | No | ||
| 4 | NUP160 | 327 | 9.292 | 0.0444 | No | ||
| 5 | TXNIP | 330 | 9.265 | 0.0534 | No | ||
| 6 | HNRPAB | 373 | 8.930 | 0.0603 | No | ||
| 7 | SRRM1 | 375 | 8.917 | 0.0690 | No | ||
| 8 | TUBA1 | 511 | 8.088 | 0.0707 | No | ||
| 9 | LIMK2 | 596 | 7.693 | 0.0743 | No | ||
| 10 | PPP1R10 | 600 | 7.677 | 0.0817 | No | ||
| 11 | SFPQ | 633 | 7.555 | 0.0877 | No | ||
| 12 | LDHB | 657 | 7.455 | 0.0939 | No | ||
| 13 | TGFB1 | 684 | 7.346 | 0.0999 | No | ||
| 14 | SF1 | 922 | 6.531 | 0.0954 | No | ||
| 15 | MAP3K11 | 963 | 6.423 | 0.0999 | No | ||
| 16 | SNRP70 | 984 | 6.370 | 0.1052 | No | ||
| 17 | PFKP | 1056 | 6.197 | 0.1080 | No | ||
| 18 | PRKCBP1 | 1135 | 6.012 | 0.1103 | No | ||
| 19 | PFN1 | 1183 | 5.900 | 0.1139 | No | ||
| 20 | RBM14 | 1268 | 5.700 | 0.1156 | No | ||
| 21 | IMPDH1 | 1352 | 5.503 | 0.1172 | No | ||
| 22 | MAZ | 1363 | 5.461 | 0.1221 | No | ||
| 23 | CLIC1 | 1566 | 5.097 | 0.1178 | No | ||
| 24 | YPEL1 | 1606 | 5.036 | 0.1210 | No | ||
| 25 | CHD4 | 1814 | 4.734 | 0.1161 | No | ||
| 26 | MYST1 | 1929 | 4.564 | 0.1153 | No | ||
| 27 | DYRK1A | 2032 | 4.420 | 0.1149 | No | ||
| 28 | DNAJB6 | 2153 | 4.274 | 0.1136 | No | ||
| 29 | SACS | 2194 | 4.229 | 0.1159 | No | ||
| 30 | FUS | 2229 | 4.200 | 0.1184 | No | ||
| 31 | BCAP31 | 2330 | 4.084 | 0.1178 | No | ||
| 32 | DHCR24 | 2610 | 3.802 | 0.1087 | No | ||
| 33 | GSPT1 | 2674 | 3.729 | 0.1095 | No | ||
| 34 | CELSR2 | 2696 | 3.707 | 0.1121 | No | ||
| 35 | GOLGA1 | 2812 | 3.600 | 0.1104 | No | ||
| 36 | COG2 | 2915 | 3.505 | 0.1091 | No | ||
| 37 | ADCY3 | 2916 | 3.504 | 0.1126 | No | ||
| 38 | TAP1 | 2968 | 3.462 | 0.1136 | No | ||
| 39 | HNRPL | 3027 | 3.413 | 0.1143 | No | ||
| 40 | PCBP1 | 3050 | 3.396 | 0.1166 | No | ||
| 41 | PABPN1 | 3086 | 3.354 | 0.1183 | No | ||
| 42 | CDC20 | 3205 | 3.256 | 0.1160 | No | ||
| 43 | DHCR7 | 3291 | 3.184 | 0.1152 | No | ||
| 44 | ME2 | 3295 | 3.178 | 0.1182 | No | ||
| 45 | UBE2M | 3308 | 3.171 | 0.1208 | No | ||
| 46 | MPHOSPH10 | 3355 | 3.132 | 0.1217 | No | ||
| 47 | FDFT1 | 3414 | 3.094 | 0.1221 | No | ||
| 48 | SSB | 3570 | 2.967 | 0.1179 | No | ||
| 49 | STAT1 | 3835 | 2.777 | 0.1084 | No | ||
| 50 | OGFR | 4070 | 2.607 | 0.1002 | No | ||
| 51 | FKBP4 | 4279 | 2.454 | 0.0930 | No | ||
| 52 | MMP15 | 4281 | 2.453 | 0.0954 | No | ||
| 53 | MRPL23 | 4443 | 2.358 | 0.0903 | No | ||
| 54 | PTPN6 | 4545 | 2.295 | 0.0879 | No | ||
| 55 | RBBP6 | 4707 | 2.195 | 0.0826 | No | ||
| 56 | TOP1 | 4795 | 2.149 | 0.0807 | No | ||
| 57 | UBE2G2 | 5005 | 2.031 | 0.0731 | No | ||
| 58 | SGTA | 5240 | 1.906 | 0.0642 | No | ||
| 59 | RPLP2 | 5445 | 1.811 | 0.0566 | No | ||
| 60 | IDH1 | 6019 | 1.533 | 0.0317 | No | ||
| 61 | KHSRP | 6077 | 1.509 | 0.0305 | No | ||
| 62 | CCNB1 | 6128 | 1.487 | 0.0297 | No | ||
| 63 | DOK4 | 6206 | 1.446 | 0.0276 | No | ||
| 64 | PSME4 | 6224 | 1.439 | 0.0282 | No | ||
| 65 | MLF2 | 6279 | 1.415 | 0.0271 | No | ||
| 66 | PSMB6 | 6284 | 1.412 | 0.0283 | No | ||
| 67 | FKBP2 | 6598 | 1.258 | 0.0151 | No | ||
| 68 | TUBG1 | 7124 | 1.028 | -0.0081 | No | ||
| 69 | EIF2B5 | 7335 | 0.936 | -0.0168 | No | ||
| 70 | GCDH | 7381 | 0.917 | -0.0180 | No | ||
| 71 | ZMPSTE24 | 7458 | 0.885 | -0.0206 | No | ||
| 72 | NDUFS6 | 9019 | 0.226 | -0.0922 | No | ||
| 73 | EMP3 | 9212 | 0.142 | -0.1009 | No | ||
| 74 | EIF2B4 | 9387 | 0.064 | -0.1089 | No | ||
| 75 | PDCD4 | 9749 | -0.108 | -0.1254 | No | ||
| 76 | ALG8 | 9796 | -0.127 | -0.1274 | No | ||
| 77 | PSMB3 | 10201 | -0.297 | -0.1457 | No | ||
| 78 | ACAT2 | 10203 | -0.297 | -0.1455 | No | ||
| 79 | ARS2 | 10216 | -0.302 | -0.1457 | No | ||
| 80 | MYD88 | 10348 | -0.356 | -0.1514 | No | ||
| 81 | POLR2C | 10515 | -0.411 | -0.1587 | No | ||
| 82 | ATP5G3 | 10692 | -0.472 | -0.1663 | No | ||
| 83 | PPP2R2A | 11150 | -0.638 | -0.1867 | No | ||
| 84 | PWP2H | 11442 | -0.730 | -0.1994 | No | ||
| 85 | SURF5 | 11712 | -0.830 | -0.2110 | No | ||
| 86 | BTK | 12025 | -0.929 | -0.2244 | No | ||
| 87 | SLC4A7 | 12054 | -0.937 | -0.2248 | No | ||
| 88 | GJB1 | 12095 | -0.948 | -0.2257 | No | ||
| 89 | LAMB3 | 12349 | -1.021 | -0.2364 | No | ||
| 90 | EBP | 12446 | -1.049 | -0.2398 | No | ||
| 91 | INHBC | 13326 | -1.309 | -0.2790 | No | ||
| 92 | NP | 13376 | -1.324 | -0.2799 | No | ||
| 93 | RPS26 | 13546 | -1.378 | -0.2863 | No | ||
| 94 | AHSA1 | 13811 | -1.454 | -0.2971 | No | ||
| 95 | CSE1L | 13929 | -1.485 | -0.3010 | No | ||
| 96 | EIF4G1 | 14061 | -1.526 | -0.3055 | No | ||
| 97 | PPIB | 14131 | -1.543 | -0.3072 | No | ||
| 98 | PSMD8 | 14190 | -1.564 | -0.3083 | No | ||
| 99 | DHX9 | 14626 | -1.690 | -0.3267 | No | ||
| 100 | MRPS12 | 14711 | -1.715 | -0.3289 | No | ||
| 101 | FASN | 14913 | -1.774 | -0.3364 | No | ||
| 102 | BYSL | 15131 | -1.846 | -0.3446 | No | ||
| 103 | EWSR1 | 15336 | -1.905 | -0.3521 | No | ||
| 104 | GRIN2A | 15580 | -1.993 | -0.3614 | No | ||
| 105 | ATR | 15642 | -2.012 | -0.3622 | No | ||
| 106 | TARDBP | 15658 | -2.018 | -0.3609 | No | ||
| 107 | PSMD3 | 15684 | -2.029 | -0.3601 | No | ||
| 108 | ACLY | 15707 | -2.035 | -0.3591 | No | ||
| 109 | ILF2 | 15849 | -2.078 | -0.3636 | No | ||
| 110 | HSPE1 | 15969 | -2.115 | -0.3670 | No | ||
| 111 | SEC61B | 15974 | -2.117 | -0.3651 | No | ||
| 112 | CCND1 | 16273 | -2.210 | -0.3766 | No | ||
| 113 | TCEB1 | 16349 | -2.234 | -0.3779 | No | ||
| 114 | RNASEH1 | 16361 | -2.237 | -0.3762 | No | ||
| 115 | IFI30 | 16382 | -2.243 | -0.3749 | No | ||
| 116 | SLC5A6 | 16477 | -2.283 | -0.3770 | No | ||
| 117 | ADAM19 | 16565 | -2.312 | -0.3788 | No | ||
| 118 | DHX15 | 16890 | -2.427 | -0.3913 | No | ||
| 119 | EIF4G2 | 16984 | -2.461 | -0.3932 | No | ||
| 120 | HSPA5 | 17144 | -2.514 | -0.3980 | No | ||
| 121 | DDX48 | 17317 | -2.582 | -0.4034 | No | ||
| 122 | EIF5 | 17444 | -2.630 | -0.4066 | No | ||
| 123 | ALG3 | 17559 | -2.665 | -0.4093 | No | ||
| 124 | IDI1 | 17723 | -2.738 | -0.4141 | No | ||
| 125 | TBL3 | 17726 | -2.739 | -0.4115 | No | ||
| 126 | SFRS1 | 17733 | -2.742 | -0.4091 | No | ||
| 127 | BMS1L | 17803 | -2.775 | -0.4095 | No | ||
| 128 | CYC1 | 17827 | -2.784 | -0.4079 | No | ||
| 129 | HRAS | 18182 | -2.929 | -0.4213 | No | ||
| 130 | PRPF4 | 18219 | -2.942 | -0.4201 | No | ||
| 131 | RUVBL2 | 18250 | -2.955 | -0.4186 | No | ||
| 132 | SMARCC1 | 18284 | -2.969 | -0.4172 | No | ||
| 133 | RABGGTB | 18362 | -3.006 | -0.4178 | No | ||
| 134 | SRPK1 | 18522 | -3.083 | -0.4221 | No | ||
| 135 | MRPL28 | 18588 | -3.115 | -0.4220 | No | ||
| 136 | SNRPF | 18641 | -3.146 | -0.4213 | No | ||
| 137 | BAZ1A | 18669 | -3.160 | -0.4195 | No | ||
| 138 | SSBP1 | 18821 | -3.245 | -0.4232 | Yes | ||
| 139 | RRS1 | 18837 | -3.257 | -0.4207 | Yes | ||
| 140 | EIF5A | 18853 | -3.265 | -0.4182 | Yes | ||
| 141 | SYNCRIP | 18869 | -3.273 | -0.4157 | Yes | ||
| 142 | PIK3R2 | 19008 | -3.358 | -0.4188 | Yes | ||
| 143 | LMNB1 | 19119 | -3.431 | -0.4205 | Yes | ||
| 144 | RANGAP1 | 19139 | -3.443 | -0.4180 | Yes | ||
| 145 | RNU3IP2 | 19158 | -3.456 | -0.4154 | Yes | ||
| 146 | XPO1 | 19211 | -3.485 | -0.4144 | Yes | ||
| 147 | CALR | 19271 | -3.520 | -0.4136 | Yes | ||
| 148 | ATP1B3 | 19291 | -3.533 | -0.4111 | Yes | ||
| 149 | NUP98 | 19385 | -3.602 | -0.4118 | Yes | ||
| 150 | SNRPB2 | 19426 | -3.633 | -0.4101 | Yes | ||
| 151 | PIK3R3 | 19528 | -3.708 | -0.4111 | Yes | ||
| 152 | SC4MOL | 19563 | -3.736 | -0.4090 | Yes | ||
| 153 | NOL1 | 19659 | -3.816 | -0.4096 | Yes | ||
| 154 | SCD | 19662 | -3.819 | -0.4060 | Yes | ||
| 155 | MORF4L2 | 19680 | -3.832 | -0.4030 | Yes | ||
| 156 | ERF | 19734 | -3.887 | -0.4016 | Yes | ||
| 157 | ARMET | 19739 | -3.891 | -0.3980 | Yes | ||
| 158 | HNRPA2B1 | 19749 | -3.899 | -0.3946 | Yes | ||
| 159 | SLC19A1 | 19863 | -4.005 | -0.3959 | Yes | ||
| 160 | RNF126 | 19937 | -4.066 | -0.3953 | Yes | ||
| 161 | SLC29A2 | 19964 | -4.089 | -0.3924 | Yes | ||
| 162 | BZW1 | 19965 | -4.089 | -0.3884 | Yes | ||
| 163 | SQLE | 19992 | -4.126 | -0.3856 | Yes | ||
| 164 | PTDSS1 | 20056 | -4.198 | -0.3844 | Yes | ||
| 165 | BOP1 | 20062 | -4.203 | -0.3805 | Yes | ||
| 166 | CCT2 | 20075 | -4.210 | -0.3769 | Yes | ||
| 167 | DNAJA1 | 20208 | -4.367 | -0.3787 | Yes | ||
| 168 | FBL | 20264 | -4.415 | -0.3769 | Yes | ||
| 169 | CCT6A | 20314 | -4.466 | -0.3748 | Yes | ||
| 170 | PSMD1 | 20350 | -4.509 | -0.3720 | Yes | ||
| 171 | NUDC | 20381 | -4.549 | -0.3689 | Yes | ||
| 172 | NMT1 | 20406 | -4.591 | -0.3655 | Yes | ||
| 173 | PRDX1 | 20439 | -4.644 | -0.3624 | Yes | ||
| 174 | EEF1E1 | 20455 | -4.662 | -0.3585 | Yes | ||
| 175 | UBE2N | 20532 | -4.758 | -0.3574 | Yes | ||
| 176 | PSMA6 | 20539 | -4.766 | -0.3530 | Yes | ||
| 177 | AK2 | 20581 | -4.814 | -0.3501 | Yes | ||
| 178 | RQCD1 | 20633 | -4.886 | -0.3477 | Yes | ||
| 179 | CYCS | 20634 | -4.887 | -0.3429 | Yes | ||
| 180 | ADRM1 | 20665 | -4.939 | -0.3394 | Yes | ||
| 181 | CDC25A | 20686 | -4.987 | -0.3355 | Yes | ||
| 182 | PAICS | 20688 | -4.990 | -0.3306 | Yes | ||
| 183 | UMPS | 20701 | -5.030 | -0.3262 | Yes | ||
| 184 | MRPL12 | 20741 | -5.100 | -0.3230 | Yes | ||
| 185 | IFRD2 | 20755 | -5.117 | -0.3186 | Yes | ||
| 186 | ERH | 20797 | -5.190 | -0.3154 | Yes | ||
| 187 | CCT5 | 20836 | -5.245 | -0.3120 | Yes | ||
| 188 | COPS3 | 20863 | -5.276 | -0.3080 | Yes | ||
| 189 | PSMD12 | 20898 | -5.320 | -0.3044 | Yes | ||
| 190 | TRIP13 | 20952 | -5.396 | -0.3015 | Yes | ||
| 191 | TFRC | 20958 | -5.404 | -0.2965 | Yes | ||
| 192 | HPRT1 | 20997 | -5.477 | -0.2929 | Yes | ||
| 193 | VDAC1 | 21066 | -5.638 | -0.2905 | Yes | ||
| 194 | HSPA4 | 21067 | -5.641 | -0.2849 | Yes | ||
| 195 | ALDOA | 21071 | -5.645 | -0.2795 | Yes | ||
| 196 | SRM | 21080 | -5.666 | -0.2743 | Yes | ||
| 197 | MTHFD1 | 21089 | -5.684 | -0.2691 | Yes | ||
| 198 | SYNGR2 | 21231 | -6.083 | -0.2697 | Yes | ||
| 199 | PPIF | 21245 | -6.125 | -0.2643 | Yes | ||
| 200 | PSMF1 | 21256 | -6.155 | -0.2587 | Yes | ||
| 201 | TCP1 | 21257 | -6.160 | -0.2526 | Yes | ||
| 202 | DDX1 | 21289 | -6.261 | -0.2479 | Yes | ||
| 203 | PSMD11 | 21321 | -6.341 | -0.2431 | Yes | ||
| 204 | ATP2A2 | 21331 | -6.383 | -0.2373 | Yes | ||
| 205 | NME1 | 21334 | -6.394 | -0.2311 | Yes | ||
| 206 | PRPS1 | 21340 | -6.411 | -0.2251 | Yes | ||
| 207 | DDX18 | 21372 | -6.502 | -0.2201 | Yes | ||
| 208 | TIMM17A | 21399 | -6.579 | -0.2149 | Yes | ||
| 209 | PHB | 21449 | -6.759 | -0.2105 | Yes | ||
| 210 | CTSC | 21486 | -6.873 | -0.2054 | Yes | ||
| 211 | METTL1 | 21576 | -7.228 | -0.2024 | Yes | ||
| 212 | PES1 | 21591 | -7.294 | -0.1959 | Yes | ||
| 213 | NCL | 21602 | -7.341 | -0.1892 | Yes | ||
| 214 | ABCF1 | 21606 | -7.360 | -0.1821 | Yes | ||
| 215 | AMD1 | 21631 | -7.566 | -0.1758 | Yes | ||
| 216 | SLC29A1 | 21634 | -7.574 | -0.1684 | Yes | ||
| 217 | MRPL3 | 21642 | -7.624 | -0.1613 | Yes | ||
| 218 | CTPS | 21657 | -7.720 | -0.1544 | Yes | ||
| 219 | SFRS2 | 21702 | -8.098 | -0.1485 | Yes | ||
| 220 | TOMM40 | 21709 | -8.157 | -0.1407 | Yes | ||
| 221 | GMPS | 21715 | -8.181 | -0.1329 | Yes | ||
| 222 | PA2G4 | 21722 | -8.235 | -0.1251 | Yes | ||
| 223 | PRDX4 | 21727 | -8.270 | -0.1172 | Yes | ||
| 224 | EBNA1BP2 | 21747 | -8.506 | -0.1097 | Yes | ||
| 225 | IDH3A | 21783 | -8.933 | -0.1026 | Yes | ||
| 226 | DDX21 | 21826 | -9.585 | -0.0951 | Yes | ||
| 227 | XRCC5 | 21831 | -9.700 | -0.0858 | Yes | ||
| 228 | NOL5A | 21859 | -10.573 | -0.0767 | Yes | ||
| 229 | SNRPA1 | 21866 | -10.806 | -0.0664 | Yes | ||
| 230 | MAPRE2 | 21900 | -12.093 | -0.0560 | Yes | ||
| 231 | LDHA | 21903 | -12.119 | -0.0442 | Yes | ||
| 232 | NOLC1 | 21924 | -13.825 | -0.0316 | Yes | ||
| 233 | C1QBP | 21938 | -16.533 | -0.0160 | Yes | ||
| 234 | CAD | 21940 | -16.671 | 0.0003 | Yes |
