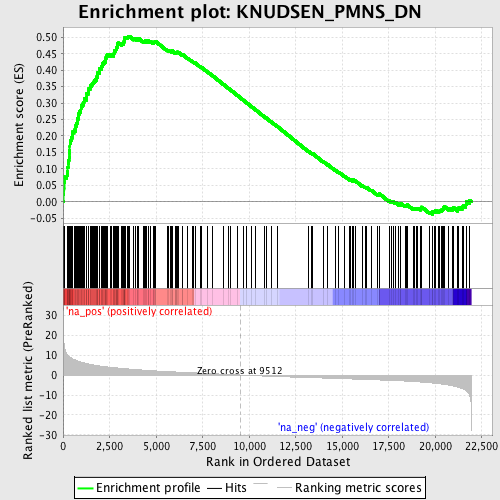
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | KNUDSEN_PMNS_DN |
| Enrichment Score (ES) | 0.5035521 |
| Normalized Enrichment Score (NES) | 2.292576 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0010064585 |
| FWER p-Value | 0.0010 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CDKN2D | 15 | 20.092 | 0.0236 | Yes | ||
| 2 | CORO1A | 47 | 15.753 | 0.0413 | Yes | ||
| 3 | ITPKB | 48 | 15.738 | 0.0604 | Yes | ||
| 4 | ARHGEF1 | 76 | 13.618 | 0.0756 | Yes | ||
| 5 | MARK2 | 208 | 10.483 | 0.0823 | Yes | ||
| 6 | ADRBK1 | 242 | 10.039 | 0.0929 | Yes | ||
| 7 | FLOT2 | 253 | 9.962 | 0.1045 | Yes | ||
| 8 | RGS14 | 269 | 9.747 | 0.1156 | Yes | ||
| 9 | LTB | 289 | 9.584 | 0.1264 | Yes | ||
| 10 | RBM5 | 320 | 9.345 | 0.1363 | Yes | ||
| 11 | TBCC | 339 | 9.185 | 0.1466 | Yes | ||
| 12 | TAOK3 | 348 | 9.105 | 0.1573 | Yes | ||
| 13 | ARHGDIB | 365 | 8.988 | 0.1674 | Yes | ||
| 14 | OAZ1 | 369 | 8.943 | 0.1781 | Yes | ||
| 15 | MTMR3 | 412 | 8.651 | 0.1866 | Yes | ||
| 16 | TSC22D3 | 440 | 8.479 | 0.1957 | Yes | ||
| 17 | PTK9L | 483 | 8.263 | 0.2037 | Yes | ||
| 18 | TUBA1 | 511 | 8.088 | 0.2123 | Yes | ||
| 19 | RARA | 585 | 7.736 | 0.2183 | Yes | ||
| 20 | RASGRP2 | 668 | 7.421 | 0.2235 | Yes | ||
| 21 | TGFB1 | 684 | 7.346 | 0.2317 | Yes | ||
| 22 | INPP5D | 724 | 7.159 | 0.2386 | Yes | ||
| 23 | STK24 | 758 | 7.030 | 0.2456 | Yes | ||
| 24 | GNAS | 780 | 6.945 | 0.2531 | Yes | ||
| 25 | RAF1 | 819 | 6.824 | 0.2596 | Yes | ||
| 26 | EDG6 | 835 | 6.783 | 0.2671 | Yes | ||
| 27 | ADD3 | 860 | 6.696 | 0.2741 | Yes | ||
| 28 | MAP3K3 | 939 | 6.496 | 0.2784 | Yes | ||
| 29 | PRKCB1 | 961 | 6.424 | 0.2852 | Yes | ||
| 30 | POLR2J | 972 | 6.409 | 0.2925 | Yes | ||
| 31 | LASP1 | 1034 | 6.252 | 0.2973 | Yes | ||
| 32 | KNS2 | 1107 | 6.071 | 0.3013 | Yes | ||
| 33 | ARHGAP4 | 1125 | 6.038 | 0.3078 | Yes | ||
| 34 | MBD4 | 1137 | 6.009 | 0.3146 | Yes | ||
| 35 | DOCK2 | 1262 | 5.718 | 0.3158 | Yes | ||
| 36 | ULK1 | 1272 | 5.688 | 0.3223 | Yes | ||
| 37 | NFE2 | 1274 | 5.671 | 0.3291 | Yes | ||
| 38 | CUGBP2 | 1341 | 5.526 | 0.3328 | Yes | ||
| 39 | ACVR1B | 1375 | 5.443 | 0.3379 | Yes | ||
| 40 | GTF3C1 | 1384 | 5.424 | 0.3441 | Yes | ||
| 41 | NUP214 | 1449 | 5.310 | 0.3475 | Yes | ||
| 42 | ITGB2 | 1471 | 5.262 | 0.3529 | Yes | ||
| 43 | GNAI2 | 1546 | 5.136 | 0.3558 | Yes | ||
| 44 | FKBP1A | 1567 | 5.097 | 0.3610 | Yes | ||
| 45 | IGF2R | 1633 | 4.999 | 0.3641 | Yes | ||
| 46 | RCBTB2 | 1679 | 4.927 | 0.3680 | Yes | ||
| 47 | USP15 | 1732 | 4.851 | 0.3715 | Yes | ||
| 48 | UBN1 | 1782 | 4.782 | 0.3750 | Yes | ||
| 49 | RERE | 1811 | 4.736 | 0.3795 | Yes | ||
| 50 | PPP1CC | 1837 | 4.695 | 0.3840 | Yes | ||
| 51 | PRMT2 | 1855 | 4.672 | 0.3889 | Yes | ||
| 52 | TAF4 | 1867 | 4.660 | 0.3940 | Yes | ||
| 53 | GNAQ | 1963 | 4.515 | 0.3951 | Yes | ||
| 54 | MAPK3 | 1966 | 4.511 | 0.4005 | Yes | ||
| 55 | MYO9B | 1969 | 4.507 | 0.4058 | Yes | ||
| 56 | CRADD | 2050 | 4.400 | 0.4075 | Yes | ||
| 57 | RABIF | 2073 | 4.378 | 0.4118 | Yes | ||
| 58 | FOXO1A | 2101 | 4.348 | 0.4158 | Yes | ||
| 59 | FLI1 | 2113 | 4.325 | 0.4205 | Yes | ||
| 60 | DDX23 | 2178 | 4.247 | 0.4227 | Yes | ||
| 61 | CREBBP | 2209 | 4.217 | 0.4265 | Yes | ||
| 62 | SPTLC1 | 2265 | 4.153 | 0.4290 | Yes | ||
| 63 | PHC2 | 2286 | 4.125 | 0.4330 | Yes | ||
| 64 | HCLS1 | 2300 | 4.113 | 0.4374 | Yes | ||
| 65 | PECAM1 | 2316 | 4.098 | 0.4417 | Yes | ||
| 66 | ACTN4 | 2352 | 4.063 | 0.4450 | Yes | ||
| 67 | OGT | 2385 | 4.030 | 0.4484 | Yes | ||
| 68 | HDAC5 | 2567 | 3.840 | 0.4447 | Yes | ||
| 69 | STK38 | 2602 | 3.807 | 0.4478 | Yes | ||
| 70 | TOB1 | 2714 | 3.696 | 0.4472 | Yes | ||
| 71 | ABCG1 | 2732 | 3.672 | 0.4508 | Yes | ||
| 72 | IQGAP1 | 2741 | 3.667 | 0.4549 | Yes | ||
| 73 | PPP1R12B | 2750 | 3.658 | 0.4590 | Yes | ||
| 74 | NCOA1 | 2802 | 3.608 | 0.4610 | Yes | ||
| 75 | ZFP36L2 | 2841 | 3.576 | 0.4636 | Yes | ||
| 76 | SLC23A2 | 2872 | 3.538 | 0.4665 | Yes | ||
| 77 | SRPK2 | 2883 | 3.531 | 0.4703 | Yes | ||
| 78 | RBMS1 | 2906 | 3.511 | 0.4735 | Yes | ||
| 79 | TBPL1 | 2909 | 3.510 | 0.4777 | Yes | ||
| 80 | GTF2B | 2937 | 3.485 | 0.4807 | Yes | ||
| 81 | GSK3B | 2980 | 3.451 | 0.4829 | Yes | ||
| 82 | STAT6 | 3153 | 3.303 | 0.4790 | Yes | ||
| 83 | PPM1A | 3168 | 3.292 | 0.4823 | Yes | ||
| 84 | TUBA2 | 3249 | 3.224 | 0.4826 | Yes | ||
| 85 | TOPBP1 | 3265 | 3.214 | 0.4858 | Yes | ||
| 86 | SMARCA2 | 3274 | 3.202 | 0.4893 | Yes | ||
| 87 | ME2 | 3295 | 3.178 | 0.4922 | Yes | ||
| 88 | ARHGAP1 | 3297 | 3.177 | 0.4960 | Yes | ||
| 89 | PLCB2 | 3318 | 3.167 | 0.4989 | Yes | ||
| 90 | YY1 | 3362 | 3.127 | 0.5007 | Yes | ||
| 91 | FADD | 3443 | 3.070 | 0.5008 | Yes | ||
| 92 | TADA3L | 3488 | 3.027 | 0.5024 | Yes | ||
| 93 | ST8SIA4 | 3543 | 2.982 | 0.5036 | Yes | ||
| 94 | AKT1 | 3802 | 2.803 | 0.4951 | No | ||
| 95 | PPP1R12A | 3870 | 2.745 | 0.4953 | No | ||
| 96 | GPR19 | 4003 | 2.644 | 0.4925 | No | ||
| 97 | CD97 | 4011 | 2.639 | 0.4953 | No | ||
| 98 | GNB2 | 4074 | 2.605 | 0.4956 | No | ||
| 99 | CDIPT | 4319 | 2.428 | 0.4874 | No | ||
| 100 | PPFIA1 | 4360 | 2.407 | 0.4884 | No | ||
| 101 | SFRS4 | 4403 | 2.381 | 0.4894 | No | ||
| 102 | TGFA | 4506 | 2.321 | 0.4875 | No | ||
| 103 | NUP153 | 4569 | 2.279 | 0.4874 | No | ||
| 104 | MAPKAPK3 | 4580 | 2.275 | 0.4897 | No | ||
| 105 | PRKAR1A | 4677 | 2.212 | 0.4880 | No | ||
| 106 | MLLT7 | 4844 | 2.127 | 0.4829 | No | ||
| 107 | PHKA2 | 4878 | 2.109 | 0.4839 | No | ||
| 108 | CBX1 | 4893 | 2.100 | 0.4858 | No | ||
| 109 | RALBP1 | 4938 | 2.076 | 0.4863 | No | ||
| 110 | ARID4A | 4963 | 2.060 | 0.4877 | No | ||
| 111 | RPS6KA5 | 5605 | 1.724 | 0.4603 | No | ||
| 112 | JAK1 | 5653 | 1.700 | 0.4602 | No | ||
| 113 | TPST2 | 5749 | 1.662 | 0.4579 | No | ||
| 114 | MAP2K4 | 5823 | 1.626 | 0.4565 | No | ||
| 115 | PRCP | 5853 | 1.615 | 0.4571 | No | ||
| 116 | APAF1 | 5854 | 1.615 | 0.4591 | No | ||
| 117 | TROVE2 | 6034 | 1.527 | 0.4527 | No | ||
| 118 | GNAO1 | 6037 | 1.526 | 0.4544 | No | ||
| 119 | TST | 6092 | 1.507 | 0.4538 | No | ||
| 120 | TYK2 | 6101 | 1.504 | 0.4552 | No | ||
| 121 | SMC1A | 6137 | 1.483 | 0.4554 | No | ||
| 122 | XPC | 6186 | 1.456 | 0.4550 | No | ||
| 123 | LST1 | 6389 | 1.368 | 0.4474 | No | ||
| 124 | HNRPUL1 | 6409 | 1.356 | 0.4481 | No | ||
| 125 | PIGB | 6695 | 1.213 | 0.4365 | No | ||
| 126 | WAS | 6943 | 1.105 | 0.4265 | No | ||
| 127 | PHKB | 7021 | 1.071 | 0.4242 | No | ||
| 128 | CD46 | 7089 | 1.043 | 0.4224 | No | ||
| 129 | HHEX | 7379 | 0.918 | 0.4102 | No | ||
| 130 | ZMPSTE24 | 7458 | 0.885 | 0.4077 | No | ||
| 131 | CSF3R | 7734 | 0.766 | 0.3960 | No | ||
| 132 | CSTA | 8030 | 0.648 | 0.3832 | No | ||
| 133 | IL8RB | 8605 | 0.406 | 0.3573 | No | ||
| 134 | UPF1 | 8899 | 0.285 | 0.3442 | No | ||
| 135 | ARPC3 | 8984 | 0.239 | 0.3406 | No | ||
| 136 | LBR | 9348 | 0.084 | 0.3240 | No | ||
| 137 | GALNT3 | 9695 | -0.083 | 0.3082 | No | ||
| 138 | TCAP | 9841 | -0.154 | 0.3017 | No | ||
| 139 | RALY | 10117 | -0.266 | 0.2894 | No | ||
| 140 | GCA | 10316 | -0.345 | 0.2807 | No | ||
| 141 | STX16 | 10798 | -0.512 | 0.2592 | No | ||
| 142 | RGS2 | 10906 | -0.550 | 0.2550 | No | ||
| 143 | TLR1 | 11215 | -0.660 | 0.2416 | No | ||
| 144 | DAPK2 | 11495 | -0.747 | 0.2297 | No | ||
| 145 | PNN | 13183 | -1.270 | 0.1536 | No | ||
| 146 | ADAM10 | 13366 | -1.321 | 0.1469 | No | ||
| 147 | TLR6 | 13425 | -1.340 | 0.1458 | No | ||
| 148 | PITPNM1 | 14003 | -1.508 | 0.1211 | No | ||
| 149 | UBE1C | 14223 | -1.574 | 0.1129 | No | ||
| 150 | DHX9 | 14626 | -1.690 | 0.0965 | No | ||
| 151 | CALM2 | 14820 | -1.748 | 0.0897 | No | ||
| 152 | SEC14L1 | 15129 | -1.846 | 0.0778 | No | ||
| 153 | PEX1 | 15397 | -1.924 | 0.0679 | No | ||
| 154 | FEZ2 | 15431 | -1.934 | 0.0687 | No | ||
| 155 | RIPK1 | 15573 | -1.990 | 0.0646 | No | ||
| 156 | RPS6KA1 | 15581 | -1.993 | 0.0667 | No | ||
| 157 | ACLY | 15707 | -2.035 | 0.0634 | No | ||
| 158 | SNAP23 | 16064 | -2.142 | 0.0496 | No | ||
| 159 | RGS19 | 16230 | -2.196 | 0.0447 | No | ||
| 160 | SLC31A2 | 16327 | -2.224 | 0.0430 | No | ||
| 161 | GNAT2 | 16550 | -2.307 | 0.0356 | No | ||
| 162 | RASA1 | 16898 | -2.430 | 0.0226 | No | ||
| 163 | BARX2 | 16973 | -2.456 | 0.0221 | No | ||
| 164 | EIF4G2 | 16984 | -2.461 | 0.0247 | No | ||
| 165 | GAS7 | 17519 | -2.654 | 0.0033 | No | ||
| 166 | UQCR | 17636 | -2.697 | 0.0013 | No | ||
| 167 | SFRS1 | 17733 | -2.742 | 0.0002 | No | ||
| 168 | CPNE3 | 17878 | -2.801 | -0.0031 | No | ||
| 169 | IDS | 18046 | -2.875 | -0.0073 | No | ||
| 170 | HTATIP2 | 18119 | -2.905 | -0.0071 | No | ||
| 171 | ACTN1 | 18148 | -2.914 | -0.0048 | No | ||
| 172 | CASP8 | 18380 | -3.013 | -0.0118 | No | ||
| 173 | GTF2E1 | 18445 | -3.047 | -0.0110 | No | ||
| 174 | USP1 | 18480 | -3.063 | -0.0089 | No | ||
| 175 | TNPO3 | 18814 | -3.242 | -0.0203 | No | ||
| 176 | CDC2L5 | 18880 | -3.280 | -0.0193 | No | ||
| 177 | CALM1 | 18993 | -3.349 | -0.0204 | No | ||
| 178 | PDE3B | 19067 | -3.392 | -0.0196 | No | ||
| 179 | GDI2 | 19190 | -3.477 | -0.0210 | No | ||
| 180 | SPAST | 19232 | -3.497 | -0.0187 | No | ||
| 181 | COX5B | 19264 | -3.515 | -0.0159 | No | ||
| 182 | HSBP1 | 19710 | -3.862 | -0.0317 | No | ||
| 183 | SLC19A1 | 19863 | -4.005 | -0.0338 | No | ||
| 184 | PLCG2 | 19865 | -4.009 | -0.0290 | No | ||
| 185 | MYCBP | 19976 | -4.104 | -0.0291 | No | ||
| 186 | SET | 20004 | -4.145 | -0.0253 | No | ||
| 187 | LRRFIP1 | 20190 | -4.340 | -0.0285 | No | ||
| 188 | SRP9 | 20247 | -4.399 | -0.0258 | No | ||
| 189 | TIAL1 | 20346 | -4.505 | -0.0248 | No | ||
| 190 | VIM | 20407 | -4.592 | -0.0220 | No | ||
| 191 | PSCD1 | 20422 | -4.620 | -0.0171 | No | ||
| 192 | CAPZB | 20479 | -4.699 | -0.0140 | No | ||
| 193 | PRDX3 | 20726 | -5.075 | -0.0191 | No | ||
| 194 | ANXA11 | 20904 | -5.329 | -0.0208 | No | ||
| 195 | NCF4 | 20973 | -5.432 | -0.0174 | No | ||
| 196 | CITED2 | 21215 | -6.023 | -0.0212 | No | ||
| 197 | CAMK2G | 21269 | -6.196 | -0.0161 | No | ||
| 198 | COPS5 | 21437 | -6.717 | -0.0156 | No | ||
| 199 | TKT | 21504 | -6.943 | -0.0103 | No | ||
| 200 | SLC25A3 | 21648 | -7.658 | -0.0076 | No | ||
| 201 | TRIM27 | 21673 | -7.831 | 0.0008 | No | ||
| 202 | XRCC5 | 21831 | -9.700 | 0.0053 | No |