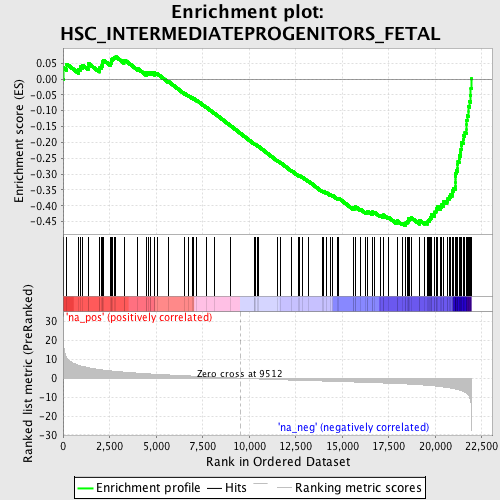
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSC_INTERMEDIATEPROGENITORS_FETAL |
| Enrichment Score (ES) | -0.46368513 |
| Normalized Enrichment Score (NES) | -2.175214 |
| Nominal p-value | 0.0 |
| FDR q-value | 9.3557197E-4 |
| FWER p-Value | 0.017 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CASP7 | 14 | 20.405 | 0.0362 | No | ||
| 2 | PYCARD | 197 | 10.649 | 0.0471 | No | ||
| 3 | CARD11 | 836 | 6.773 | 0.0301 | No | ||
| 4 | RELA | 916 | 6.549 | 0.0383 | No | ||
| 5 | PFKP | 1056 | 6.197 | 0.0431 | No | ||
| 6 | FIS1 | 1347 | 5.512 | 0.0397 | No | ||
| 7 | IMPDH1 | 1352 | 5.503 | 0.0495 | No | ||
| 8 | ISG20L2 | 1950 | 4.539 | 0.0303 | No | ||
| 9 | LMAN2 | 1975 | 4.498 | 0.0373 | No | ||
| 10 | GMPPA | 2063 | 4.387 | 0.0413 | No | ||
| 11 | PINK1 | 2103 | 4.345 | 0.0473 | No | ||
| 12 | CDADC1 | 2127 | 4.310 | 0.0540 | No | ||
| 13 | C5ORF14 | 2173 | 4.255 | 0.0597 | No | ||
| 14 | E2F8 | 2568 | 3.837 | 0.0485 | No | ||
| 15 | D4ST1 | 2584 | 3.824 | 0.0547 | No | ||
| 16 | RAPH1 | 2594 | 3.816 | 0.0612 | No | ||
| 17 | GSPT1 | 2674 | 3.729 | 0.0643 | No | ||
| 18 | FANCM | 2734 | 3.671 | 0.0683 | No | ||
| 19 | NEBL | 2840 | 3.578 | 0.0699 | No | ||
| 20 | SCIN | 3272 | 3.204 | 0.0559 | No | ||
| 21 | NUDT16L1 | 3314 | 3.169 | 0.0598 | No | ||
| 22 | AP3S2 | 4017 | 2.635 | 0.0323 | No | ||
| 23 | CUL5 | 4468 | 2.343 | 0.0159 | No | ||
| 24 | SFXN4 | 4480 | 2.332 | 0.0196 | No | ||
| 25 | MAPKAPK3 | 4580 | 2.275 | 0.0192 | No | ||
| 26 | NPAL1 | 4697 | 2.200 | 0.0179 | No | ||
| 27 | ATP6V1F | 4711 | 2.194 | 0.0212 | No | ||
| 28 | TEX2 | 4907 | 2.094 | 0.0161 | No | ||
| 29 | QARS | 4908 | 2.093 | 0.0199 | No | ||
| 30 | VPS45A | 5058 | 1.998 | 0.0166 | No | ||
| 31 | MRPS25 | 5659 | 1.698 | -0.0078 | No | ||
| 32 | WNT2 | 6534 | 1.291 | -0.0455 | No | ||
| 33 | KLHL23 | 6722 | 1.205 | -0.0519 | No | ||
| 34 | WDR73 | 6928 | 1.110 | -0.0593 | No | ||
| 35 | RASSF4 | 7030 | 1.067 | -0.0620 | No | ||
| 36 | GYS1 | 7193 | 1.002 | -0.0677 | No | ||
| 37 | AS3MT | 7681 | 0.791 | -0.0886 | No | ||
| 38 | GRHL3 | 8123 | 0.609 | -0.1077 | No | ||
| 39 | SCO1 | 8980 | 0.244 | -0.1465 | No | ||
| 40 | OVOL2 | 10302 | -0.336 | -0.2064 | No | ||
| 41 | WDR34 | 10304 | -0.337 | -0.2059 | No | ||
| 42 | PLEKHO1 | 10346 | -0.356 | -0.2071 | No | ||
| 43 | EEF1B2 | 10449 | -0.388 | -0.2111 | No | ||
| 44 | SURF6 | 10483 | -0.400 | -0.2119 | No | ||
| 45 | SDF2L1 | 11525 | -0.757 | -0.2582 | No | ||
| 46 | PLA2G4D | 11680 | -0.816 | -0.2638 | No | ||
| 47 | CPA3 | 12281 | -1.001 | -0.2895 | No | ||
| 48 | THOC5 | 12648 | -1.110 | -0.3043 | No | ||
| 49 | ELOVL3 | 12708 | -1.127 | -0.3050 | No | ||
| 50 | GIP | 12852 | -1.171 | -0.3094 | No | ||
| 51 | NOSTRIN | 13190 | -1.271 | -0.3226 | No | ||
| 52 | SLC35B2 | 13926 | -1.485 | -0.3536 | No | ||
| 53 | BET1 | 14017 | -1.514 | -0.3550 | No | ||
| 54 | XPA | 14137 | -1.545 | -0.3576 | No | ||
| 55 | KLHDC4 | 14346 | -1.609 | -0.3643 | No | ||
| 56 | GPR180 | 14471 | -1.639 | -0.3670 | No | ||
| 57 | BLOC1S2 | 14768 | -1.731 | -0.3774 | No | ||
| 58 | RPUSD3 | 14811 | -1.745 | -0.3762 | No | ||
| 59 | SPFH1 | 15597 | -1.999 | -0.4086 | No | ||
| 60 | GALK1 | 15598 | -1.999 | -0.4050 | No | ||
| 61 | DYNLL1 | 15713 | -2.036 | -0.4065 | No | ||
| 62 | ERCC8 | 15715 | -2.037 | -0.4029 | No | ||
| 63 | TNS3 | 16004 | -2.123 | -0.4123 | No | ||
| 64 | MTG1 | 16270 | -2.209 | -0.4204 | No | ||
| 65 | GTPBP8 | 16335 | -2.228 | -0.4194 | No | ||
| 66 | CEBPZ | 16363 | -2.237 | -0.4166 | No | ||
| 67 | CTSZ | 16607 | -2.324 | -0.4235 | No | ||
| 68 | THRAP3 | 16608 | -2.326 | -0.4193 | No | ||
| 69 | STATIP1 | 16735 | -2.370 | -0.4208 | No | ||
| 70 | DDX52 | 17054 | -2.484 | -0.4309 | No | ||
| 71 | PLAA | 17207 | -2.538 | -0.4333 | No | ||
| 72 | PAPSS2 | 17211 | -2.539 | -0.4288 | No | ||
| 73 | SSR2 | 17460 | -2.634 | -0.4354 | No | ||
| 74 | ATP6AP2 | 17941 | -2.830 | -0.4523 | No | ||
| 75 | ITGB4BP | 17944 | -2.831 | -0.4473 | No | ||
| 76 | SPCS3 | 18232 | -2.949 | -0.4552 | No | ||
| 77 | SNUPN | 18419 | -3.033 | -0.4582 | Yes | ||
| 78 | DTX4 | 18422 | -3.035 | -0.4528 | Yes | ||
| 79 | NUDT22 | 18481 | -3.064 | -0.4499 | Yes | ||
| 80 | NUP37 | 18535 | -3.089 | -0.4468 | Yes | ||
| 81 | DDX20 | 18536 | -3.090 | -0.4412 | Yes | ||
| 82 | NTHL1 | 18625 | -3.137 | -0.4396 | Yes | ||
| 83 | THOP1 | 18711 | -3.185 | -0.4377 | Yes | ||
| 84 | CLUAP1 | 19149 | -3.450 | -0.4515 | Yes | ||
| 85 | RABEPK | 19153 | -3.452 | -0.4454 | Yes | ||
| 86 | SCARB1 | 19423 | -3.632 | -0.4512 | Yes | ||
| 87 | CTNNA3 | 19565 | -3.737 | -0.4509 | Yes | ||
| 88 | DBI | 19606 | -3.775 | -0.4460 | Yes | ||
| 89 | MAOA | 19699 | -3.853 | -0.4432 | Yes | ||
| 90 | RPN1 | 19735 | -3.887 | -0.4378 | Yes | ||
| 91 | CFDP1 | 19795 | -3.941 | -0.4334 | Yes | ||
| 92 | WDFY3 | 19807 | -3.956 | -0.4268 | Yes | ||
| 93 | MDH1 | 19951 | -4.074 | -0.4260 | Yes | ||
| 94 | GCNT3 | 19974 | -4.102 | -0.4196 | Yes | ||
| 95 | PCCA | 20037 | -4.182 | -0.4149 | Yes | ||
| 96 | PSPH | 20078 | -4.217 | -0.4091 | Yes | ||
| 97 | TSPAN31 | 20120 | -4.263 | -0.4033 | Yes | ||
| 98 | C12ORF10 | 20292 | -4.439 | -0.4031 | Yes | ||
| 99 | RSAD1 | 20342 | -4.499 | -0.3972 | Yes | ||
| 100 | HDGF | 20452 | -4.658 | -0.3938 | Yes | ||
| 101 | TBRG4 | 20454 | -4.662 | -0.3854 | Yes | ||
| 102 | HES6 | 20628 | -4.880 | -0.3845 | Yes | ||
| 103 | MRPS35 | 20677 | -4.969 | -0.3778 | Yes | ||
| 104 | MRPL12 | 20741 | -5.100 | -0.3715 | Yes | ||
| 105 | AGPAT5 | 20814 | -5.212 | -0.3653 | Yes | ||
| 106 | SDC4 | 20901 | -5.326 | -0.3597 | Yes | ||
| 107 | CHCHD4 | 20917 | -5.349 | -0.3507 | Yes | ||
| 108 | AHCY | 20985 | -5.458 | -0.3439 | Yes | ||
| 109 | ALDOA | 21071 | -5.645 | -0.3376 | Yes | ||
| 110 | TMEM97 | 21077 | -5.657 | -0.3276 | Yes | ||
| 111 | TCN2 | 21083 | -5.674 | -0.3176 | Yes | ||
| 112 | NPC2 | 21084 | -5.675 | -0.3074 | Yes | ||
| 113 | CCDC98 | 21098 | -5.704 | -0.2977 | Yes | ||
| 114 | TXNDC4 | 21121 | -5.759 | -0.2883 | Yes | ||
| 115 | POLR3E | 21203 | -5.991 | -0.2812 | Yes | ||
| 116 | PITPNB | 21208 | -6.006 | -0.2705 | Yes | ||
| 117 | CITED2 | 21215 | -6.023 | -0.2599 | Yes | ||
| 118 | APIP | 21280 | -6.215 | -0.2516 | Yes | ||
| 119 | WDR77 | 21294 | -6.277 | -0.2409 | Yes | ||
| 120 | RSU1 | 21365 | -6.489 | -0.2324 | Yes | ||
| 121 | GPT2 | 21373 | -6.505 | -0.2210 | Yes | ||
| 122 | GFM1 | 21392 | -6.555 | -0.2100 | Yes | ||
| 123 | POLR3C | 21414 | -6.620 | -0.1990 | Yes | ||
| 124 | NAT12 | 21513 | -6.995 | -0.1908 | Yes | ||
| 125 | APEX1 | 21526 | -7.036 | -0.1787 | Yes | ||
| 126 | PES1 | 21591 | -7.294 | -0.1684 | Yes | ||
| 127 | PLAC8 | 21682 | -7.915 | -0.1583 | Yes | ||
| 128 | PARL | 21685 | -7.976 | -0.1440 | Yes | ||
| 129 | TIMM44 | 21690 | -7.999 | -0.1297 | Yes | ||
| 130 | PTCD2 | 21707 | -8.123 | -0.1158 | Yes | ||
| 131 | BID | 21795 | -9.169 | -0.1032 | Yes | ||
| 132 | GUSB | 21797 | -9.197 | -0.0867 | Yes | ||
| 133 | COMMD10 | 21845 | -10.075 | -0.0706 | Yes | ||
| 134 | MAPRE2 | 21900 | -12.093 | -0.0513 | Yes | ||
| 135 | GTF3A | 21913 | -12.862 | -0.0286 | Yes | ||
| 136 | CAD | 21940 | -16.671 | 0.0003 | Yes |
