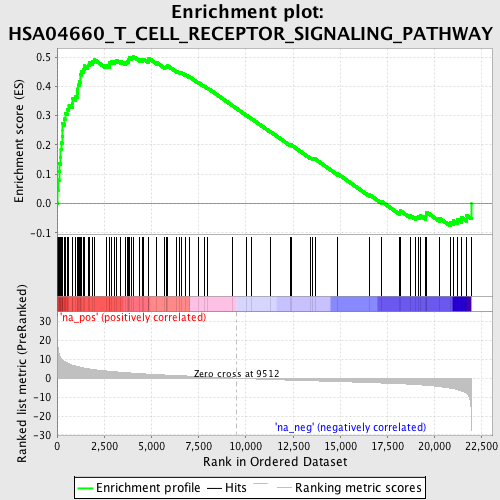
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA04660_T_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.5010915 |
| Normalized Enrichment Score (NES) | 2.050325 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0088058 |
| FWER p-Value | 0.093 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LCK | 20 | 19.234 | 0.0447 | Yes | ||
| 2 | CD3E | 51 | 15.588 | 0.0803 | Yes | ||
| 3 | TEC | 104 | 12.753 | 0.1082 | Yes | ||
| 4 | NFKBIA | 116 | 12.486 | 0.1373 | Yes | ||
| 5 | CD247 | 200 | 10.587 | 0.1587 | Yes | ||
| 6 | CD28 | 204 | 10.529 | 0.1835 | Yes | ||
| 7 | PIK3CD | 211 | 10.436 | 0.2080 | Yes | ||
| 8 | CD8A | 263 | 9.816 | 0.2290 | Yes | ||
| 9 | CD8B | 295 | 9.553 | 0.2502 | Yes | ||
| 10 | CD4 | 297 | 9.537 | 0.2728 | Yes | ||
| 11 | PIK3R5 | 401 | 8.752 | 0.2888 | Yes | ||
| 12 | VAV2 | 421 | 8.556 | 0.3083 | Yes | ||
| 13 | PLCG1 | 539 | 7.963 | 0.3218 | Yes | ||
| 14 | VAV1 | 628 | 7.582 | 0.3358 | Yes | ||
| 15 | NFATC1 | 826 | 6.800 | 0.3429 | Yes | ||
| 16 | CARD11 | 836 | 6.773 | 0.3586 | Yes | ||
| 17 | PPP3CB | 991 | 6.360 | 0.3666 | Yes | ||
| 18 | CD3D | 1059 | 6.186 | 0.3782 | Yes | ||
| 19 | ITK | 1091 | 6.101 | 0.3913 | Yes | ||
| 20 | IKBKG | 1106 | 6.072 | 0.4050 | Yes | ||
| 21 | NFATC3 | 1178 | 5.913 | 0.4158 | Yes | ||
| 22 | CBL | 1220 | 5.822 | 0.4278 | Yes | ||
| 23 | PAK4 | 1221 | 5.822 | 0.4416 | Yes | ||
| 24 | AKT2 | 1306 | 5.599 | 0.4510 | Yes | ||
| 25 | LCP2 | 1403 | 5.391 | 0.4594 | Yes | ||
| 26 | NFKB2 | 1444 | 5.315 | 0.4702 | Yes | ||
| 27 | NFAT5 | 1636 | 4.993 | 0.4733 | Yes | ||
| 28 | ICOS | 1708 | 4.885 | 0.4816 | Yes | ||
| 29 | CBLB | 1885 | 4.615 | 0.4845 | Yes | ||
| 30 | MAP3K14 | 1956 | 4.529 | 0.4921 | Yes | ||
| 31 | JUN | 2591 | 3.817 | 0.4721 | Yes | ||
| 32 | MALT1 | 2770 | 3.638 | 0.4726 | Yes | ||
| 33 | BCL10 | 2773 | 3.637 | 0.4812 | Yes | ||
| 34 | PIK3CA | 2870 | 3.541 | 0.4852 | Yes | ||
| 35 | PTPRC | 3046 | 3.398 | 0.4852 | Yes | ||
| 36 | ZAP70 | 3141 | 3.317 | 0.4888 | Yes | ||
| 37 | SOS1 | 3360 | 3.127 | 0.4862 | Yes | ||
| 38 | RASGRP1 | 3641 | 2.914 | 0.4804 | Yes | ||
| 39 | PAK1 | 3703 | 2.873 | 0.4844 | Yes | ||
| 40 | CTLA4 | 3798 | 2.805 | 0.4867 | Yes | ||
| 41 | AKT1 | 3802 | 2.803 | 0.4932 | Yes | ||
| 42 | PDK1 | 3851 | 2.765 | 0.4976 | Yes | ||
| 43 | AKT3 | 3957 | 2.670 | 0.4991 | Yes | ||
| 44 | CD3G | 4051 | 2.614 | 0.5011 | Yes | ||
| 45 | NFKBIB | 4385 | 2.393 | 0.4915 | No | ||
| 46 | PTPN6 | 4545 | 2.295 | 0.4897 | No | ||
| 47 | NCK1 | 4581 | 2.272 | 0.4935 | No | ||
| 48 | PPP3R1 | 4827 | 2.135 | 0.4874 | No | ||
| 49 | PIK3CG | 4859 | 2.117 | 0.4910 | No | ||
| 50 | PAK6 | 4864 | 2.116 | 0.4958 | No | ||
| 51 | NFKBIE | 5291 | 1.879 | 0.4808 | No | ||
| 52 | PIK3CB | 5715 | 1.678 | 0.4654 | No | ||
| 53 | PIK3R1 | 5770 | 1.654 | 0.4668 | No | ||
| 54 | GRAP2 | 5821 | 1.627 | 0.4684 | No | ||
| 55 | CSF2 | 5829 | 1.624 | 0.4720 | No | ||
| 56 | NRAS | 6347 | 1.384 | 0.4516 | No | ||
| 57 | PRKCQ | 6476 | 1.323 | 0.4489 | No | ||
| 58 | FYN | 6571 | 1.275 | 0.4476 | No | ||
| 59 | FOS | 6777 | 1.183 | 0.4410 | No | ||
| 60 | CHUK | 6991 | 1.084 | 0.4338 | No | ||
| 61 | IL4 | 7504 | 0.867 | 0.4125 | No | ||
| 62 | SOS2 | 7785 | 0.747 | 0.4014 | No | ||
| 63 | RHOA | 7991 | 0.666 | 0.3936 | No | ||
| 64 | LAT | 9292 | 0.110 | 0.3344 | No | ||
| 65 | MAP3K8 | 10010 | -0.225 | 0.3022 | No | ||
| 66 | PAK2 | 10297 | -0.335 | 0.2899 | No | ||
| 67 | CD40LG | 11288 | -0.678 | 0.2462 | No | ||
| 68 | CBLC | 12376 | -1.030 | 0.1989 | No | ||
| 69 | VAV3 | 12393 | -1.035 | 0.2006 | No | ||
| 70 | TNF | 13449 | -1.346 | 0.1555 | No | ||
| 71 | PAK7 | 13554 | -1.381 | 0.1541 | No | ||
| 72 | PPP3R2 | 13667 | -1.414 | 0.1523 | No | ||
| 73 | IL10 | 14877 | -1.762 | 0.1012 | No | ||
| 74 | IFNG | 16549 | -2.307 | 0.0302 | No | ||
| 75 | PDCD1 | 17180 | -2.525 | 0.0073 | No | ||
| 76 | NFATC4 | 18154 | -2.918 | -0.0302 | No | ||
| 77 | HRAS | 18182 | -2.929 | -0.0245 | No | ||
| 78 | PPP3CA | 18699 | -3.178 | -0.0406 | No | ||
| 79 | PIK3R2 | 19008 | -3.358 | -0.0467 | No | ||
| 80 | CDC42 | 19144 | -3.447 | -0.0447 | No | ||
| 81 | PAK3 | 19253 | -3.511 | -0.0413 | No | ||
| 82 | PIK3R3 | 19528 | -3.708 | -0.0451 | No | ||
| 83 | NFATC2 | 19548 | -3.725 | -0.0371 | No | ||
| 84 | IKBKB | 19586 | -3.755 | -0.0299 | No | ||
| 85 | KRAS | 20270 | -4.421 | -0.0506 | No | ||
| 86 | GRB2 | 20842 | -5.251 | -0.0643 | No | ||
| 87 | CDK4 | 21020 | -5.528 | -0.0593 | No | ||
| 88 | NFKB1 | 21220 | -6.043 | -0.0541 | No | ||
| 89 | PPP3CC | 21439 | -6.721 | -0.0481 | No | ||
| 90 | NCK2 | 21710 | -8.166 | -0.0411 | No | ||
| 91 | IL5 | 21945 | -21.856 | 0.0001 | No |