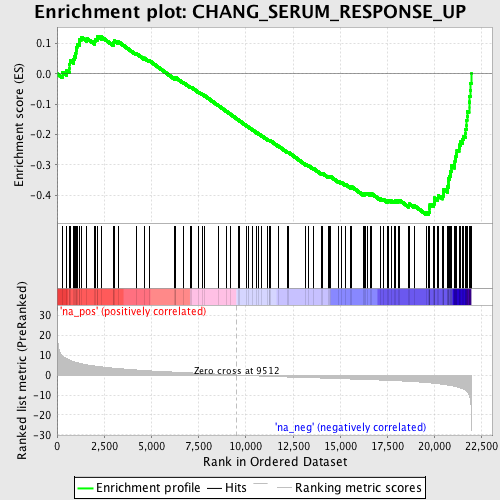
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CHANG_SERUM_RESPONSE_UP |
| Enrichment Score (ES) | -0.46299115 |
| Normalized Enrichment Score (NES) | -2.1917527 |
| Nominal p-value | 0.0 |
| FDR q-value | 7.4476056E-4 |
| FWER p-Value | 0.012 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RBMX | 267 | 9.771 | 0.0060 | No | ||
| 2 | TUBA1 | 511 | 8.088 | 0.0099 | No | ||
| 3 | DCK | 640 | 7.530 | 0.0180 | No | ||
| 4 | RNF41 | 647 | 7.518 | 0.0317 | No | ||
| 5 | CDCA4 | 682 | 7.353 | 0.0439 | No | ||
| 6 | H2AFZ | 874 | 6.647 | 0.0475 | No | ||
| 7 | VIL2 | 911 | 6.561 | 0.0581 | No | ||
| 8 | ID3 | 981 | 6.380 | 0.0668 | No | ||
| 9 | SFRS10 | 1002 | 6.335 | 0.0777 | No | ||
| 10 | CENPJ | 1037 | 6.248 | 0.0877 | No | ||
| 11 | PFKP | 1056 | 6.197 | 0.0984 | No | ||
| 12 | PFN1 | 1183 | 5.900 | 0.1036 | No | ||
| 13 | CKLF | 1200 | 5.856 | 0.1138 | No | ||
| 14 | RBM14 | 1268 | 5.700 | 0.1214 | No | ||
| 15 | COTL1 | 1580 | 5.079 | 0.1166 | No | ||
| 16 | SNRPB | 1987 | 4.478 | 0.1063 | No | ||
| 17 | PCSK7 | 2013 | 4.440 | 0.1134 | No | ||
| 18 | CDK2 | 2120 | 4.313 | 0.1166 | No | ||
| 19 | COPS6 | 2125 | 4.311 | 0.1244 | No | ||
| 20 | NUDT1 | 2344 | 4.073 | 0.1220 | No | ||
| 21 | WSB2 | 3001 | 3.433 | 0.0983 | No | ||
| 22 | ITGA6 | 3003 | 3.432 | 0.1047 | No | ||
| 23 | HNRPR | 3052 | 3.395 | 0.1088 | No | ||
| 24 | DLEU2 | 3228 | 3.240 | 0.1068 | No | ||
| 25 | PITPNC1 | 4201 | 2.506 | 0.0669 | No | ||
| 26 | MYL6 | 4609 | 2.256 | 0.0524 | No | ||
| 27 | CBX1 | 4893 | 2.100 | 0.0434 | No | ||
| 28 | HMGN2 | 6245 | 1.428 | -0.0159 | No | ||
| 29 | TNFRSF12A | 6258 | 1.423 | -0.0138 | No | ||
| 30 | SMURF2 | 6267 | 1.421 | -0.0115 | No | ||
| 31 | SNRPE | 6683 | 1.219 | -0.0283 | No | ||
| 32 | UAP1 | 7071 | 1.052 | -0.0441 | No | ||
| 33 | TUBG1 | 7124 | 1.028 | -0.0445 | No | ||
| 34 | COX17 | 7496 | 0.870 | -0.0599 | No | ||
| 35 | MRPS16 | 7683 | 0.791 | -0.0670 | No | ||
| 36 | PLOD2 | 7835 | 0.724 | -0.0725 | No | ||
| 37 | PLAUR | 8554 | 0.431 | -0.1047 | No | ||
| 38 | MET | 8955 | 0.252 | -0.1225 | No | ||
| 39 | MT3 | 9211 | 0.144 | -0.1339 | No | ||
| 40 | GPLD1 | 9600 | -0.045 | -0.1517 | No | ||
| 41 | PSMD2 | 9671 | -0.075 | -0.1547 | No | ||
| 42 | MAP3K8 | 10010 | -0.225 | -0.1698 | No | ||
| 43 | PTS | 10164 | -0.285 | -0.1763 | No | ||
| 44 | F3 | 10353 | -0.358 | -0.1842 | No | ||
| 45 | MCM3 | 10591 | -0.436 | -0.1943 | No | ||
| 46 | LSM4 | 10676 | -0.466 | -0.1973 | No | ||
| 47 | GNG11 | 10850 | -0.530 | -0.2042 | No | ||
| 48 | MSN | 11160 | -0.642 | -0.2172 | No | ||
| 49 | ADAMTS1 | 11237 | -0.666 | -0.2194 | No | ||
| 50 | BRIP1 | 11262 | -0.673 | -0.2193 | No | ||
| 51 | PLG | 11319 | -0.688 | -0.2206 | No | ||
| 52 | MKKS | 11720 | -0.833 | -0.2373 | No | ||
| 53 | TFPI2 | 12212 | -0.979 | -0.2580 | No | ||
| 54 | MCM7 | 12247 | -0.991 | -0.2577 | No | ||
| 55 | PNN | 13183 | -1.270 | -0.2982 | No | ||
| 56 | TAGLN | 13319 | -1.308 | -0.3020 | No | ||
| 57 | LMNB2 | 13575 | -1.387 | -0.3111 | No | ||
| 58 | EPHB1 | 13989 | -1.503 | -0.3272 | No | ||
| 59 | EIF4G1 | 14061 | -1.526 | -0.3277 | No | ||
| 60 | CHEK1 | 14360 | -1.611 | -0.3383 | No | ||
| 61 | PSMC3 | 14407 | -1.622 | -0.3374 | No | ||
| 62 | POLE2 | 14478 | -1.640 | -0.3376 | No | ||
| 63 | IMP4 | 14908 | -1.773 | -0.3539 | No | ||
| 64 | MRPL37 | 15048 | -1.814 | -0.3569 | No | ||
| 65 | SNRPA | 15282 | -1.888 | -0.3641 | No | ||
| 66 | RNASEH2A | 15562 | -1.986 | -0.3732 | No | ||
| 67 | FLNC | 15574 | -1.990 | -0.3700 | No | ||
| 68 | SSR3 | 16231 | -2.196 | -0.3960 | No | ||
| 69 | NUPL1 | 16301 | -2.219 | -0.3950 | No | ||
| 70 | TCEB1 | 16349 | -2.234 | -0.3930 | No | ||
| 71 | HMGB1 | 16431 | -2.263 | -0.3925 | No | ||
| 72 | HAS2 | 16601 | -2.323 | -0.3959 | No | ||
| 73 | SMS | 16654 | -2.338 | -0.3939 | No | ||
| 74 | MYBL2 | 17135 | -2.511 | -0.4113 | No | ||
| 75 | ENO1 | 17283 | -2.566 | -0.4132 | No | ||
| 76 | LSM3 | 17493 | -2.643 | -0.4179 | No | ||
| 77 | EMP2 | 17566 | -2.666 | -0.4162 | No | ||
| 78 | GGH | 17724 | -2.738 | -0.4183 | No | ||
| 79 | SLC16A1 | 17872 | -2.798 | -0.4199 | No | ||
| 80 | NUTF2 | 17912 | -2.817 | -0.4164 | No | ||
| 81 | NUP107 | 18068 | -2.885 | -0.4181 | No | ||
| 82 | EIF4EBP1 | 18133 | -2.908 | -0.4157 | No | ||
| 83 | MAPRE1 | 18636 | -3.144 | -0.4328 | No | ||
| 84 | DHFR | 18663 | -3.156 | -0.4281 | No | ||
| 85 | LOXL2 | 18929 | -3.313 | -0.4341 | No | ||
| 86 | CORO1C | 19560 | -3.735 | -0.4560 | Yes | ||
| 87 | DCLRE1B | 19702 | -3.855 | -0.4553 | Yes | ||
| 88 | RPN1 | 19735 | -3.887 | -0.4496 | Yes | ||
| 89 | RNF138 | 19736 | -3.887 | -0.4423 | Yes | ||
| 90 | HNRPA2B1 | 19749 | -3.899 | -0.4356 | Yes | ||
| 91 | DUT | 19757 | -3.904 | -0.4287 | Yes | ||
| 92 | BRCA2 | 19959 | -4.084 | -0.4303 | Yes | ||
| 93 | MYCBP | 19976 | -4.104 | -0.4234 | Yes | ||
| 94 | MNAT1 | 19983 | -4.113 | -0.4160 | Yes | ||
| 95 | RFC3 | 19986 | -4.119 | -0.4084 | Yes | ||
| 96 | MRPS28 | 20172 | -4.313 | -0.4089 | Yes | ||
| 97 | TPM2 | 20185 | -4.335 | -0.4013 | Yes | ||
| 98 | SLC25A5 | 20419 | -4.616 | -0.4034 | Yes | ||
| 99 | EEF1E1 | 20455 | -4.662 | -0.3964 | Yes | ||
| 100 | SSSCA1 | 20468 | -4.686 | -0.3882 | Yes | ||
| 101 | RUVBL1 | 20492 | -4.713 | -0.3805 | Yes | ||
| 102 | PAICS | 20688 | -4.990 | -0.3801 | Yes | ||
| 103 | UMPS | 20701 | -5.030 | -0.3713 | Yes | ||
| 104 | MRPL12 | 20741 | -5.100 | -0.3636 | Yes | ||
| 105 | JTV1 | 20747 | -5.104 | -0.3543 | Yes | ||
| 106 | IFRD2 | 20755 | -5.117 | -0.3451 | Yes | ||
| 107 | POLR3K | 20801 | -5.194 | -0.3375 | Yes | ||
| 108 | CCT5 | 20836 | -5.245 | -0.3293 | Yes | ||
| 109 | PSMA7 | 20850 | -5.266 | -0.3201 | Yes | ||
| 110 | BCCIP | 20893 | -5.316 | -0.3121 | Yes | ||
| 111 | PSMD12 | 20898 | -5.320 | -0.3024 | Yes | ||
| 112 | VDAC1 | 21066 | -5.638 | -0.2996 | Yes | ||
| 113 | SRM | 21080 | -5.666 | -0.2896 | Yes | ||
| 114 | MTHFD1 | 21089 | -5.684 | -0.2794 | Yes | ||
| 115 | IPO4 | 21131 | -5.788 | -0.2705 | Yes | ||
| 116 | PSMD14 | 21175 | -5.898 | -0.2615 | Yes | ||
| 117 | PDAP1 | 21187 | -5.915 | -0.2510 | Yes | ||
| 118 | SNRPD1 | 21298 | -6.287 | -0.2443 | Yes | ||
| 119 | NME1 | 21334 | -6.394 | -0.2340 | Yes | ||
| 120 | TXNL2 | 21378 | -6.512 | -0.2239 | Yes | ||
| 121 | NOLA2 | 21479 | -6.851 | -0.2157 | Yes | ||
| 122 | TPI1 | 21523 | -7.025 | -0.2046 | Yes | ||
| 123 | UBE2J1 | 21639 | -7.602 | -0.1957 | Yes | ||
| 124 | LYAR | 21651 | -7.674 | -0.1819 | Yes | ||
| 125 | SFRS2 | 21702 | -8.098 | -0.1691 | Yes | ||
| 126 | TOMM40 | 21709 | -8.157 | -0.1542 | Yes | ||
| 127 | PA2G4 | 21722 | -8.235 | -0.1394 | Yes | ||
| 128 | EBNA1BP2 | 21747 | -8.506 | -0.1247 | Yes | ||
| 129 | TPM1 | 21827 | -9.610 | -0.1104 | Yes | ||
| 130 | NLN | 21828 | -9.662 | -0.0924 | Yes | ||
| 131 | SNRPA1 | 21866 | -10.806 | -0.0740 | Yes | ||
| 132 | ID2 | 21890 | -11.617 | -0.0534 | Yes | ||
| 133 | IL7R | 21915 | -13.038 | -0.0303 | Yes | ||
| 134 | SDC1 | 21941 | -17.025 | 0.0003 | Yes |
