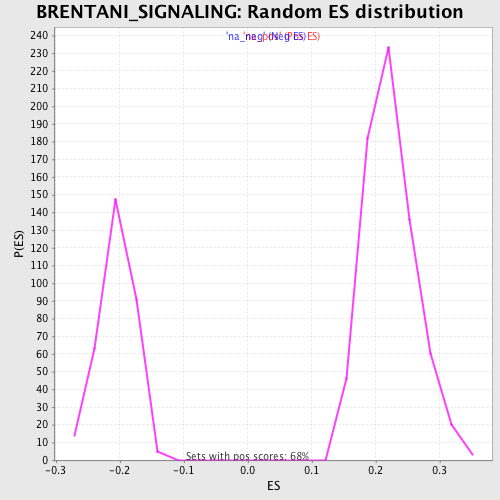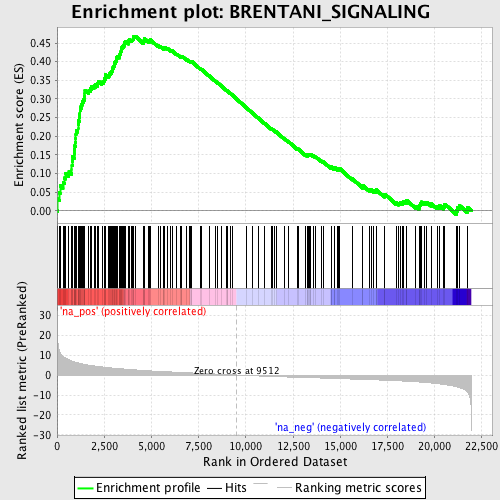
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | BRENTANI_SIGNALING |
| Enrichment Score (ES) | 0.4686606 |
| Normalized Enrichment Score (NES) | 2.1070087 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0035252771 |
| FWER p-Value | 0.027 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TNFRSF1A | 22 | 18.882 | 0.0320 | Yes | ||
| 2 | STAT5B | 125 | 12.304 | 0.0488 | Yes | ||
| 3 | PTK2B | 159 | 11.293 | 0.0671 | Yes | ||
| 4 | TRAF3 | 312 | 9.412 | 0.0765 | Yes | ||
| 5 | YWHAE | 396 | 8.835 | 0.0882 | Yes | ||
| 6 | ABL1 | 461 | 8.376 | 0.0999 | Yes | ||
| 7 | VAV1 | 628 | 7.582 | 0.1055 | Yes | ||
| 8 | IFNAR1 | 785 | 6.934 | 0.1105 | Yes | ||
| 9 | IQGAP2 | 787 | 6.930 | 0.1225 | Yes | ||
| 10 | TRAF1 | 801 | 6.898 | 0.1340 | Yes | ||
| 11 | RAF1 | 819 | 6.824 | 0.1452 | Yes | ||
| 12 | DGKG | 903 | 6.584 | 0.1529 | Yes | ||
| 13 | RELA | 916 | 6.549 | 0.1638 | Yes | ||
| 14 | MAP3K3 | 939 | 6.496 | 0.1741 | Yes | ||
| 15 | STAT2 | 975 | 6.402 | 0.1837 | Yes | ||
| 16 | MAP2K7 | 988 | 6.363 | 0.1943 | Yes | ||
| 17 | DGKQ | 996 | 6.351 | 0.2050 | Yes | ||
| 18 | MAP4K4 | 1004 | 6.331 | 0.2158 | Yes | ||
| 19 | IKBKG | 1106 | 6.072 | 0.2218 | Yes | ||
| 20 | MAP2K2 | 1109 | 6.063 | 0.2323 | Yes | ||
| 21 | IFNAR2 | 1136 | 6.010 | 0.2416 | Yes | ||
| 22 | STAT3 | 1171 | 5.916 | 0.2504 | Yes | ||
| 23 | PTEN | 1189 | 5.873 | 0.2599 | Yes | ||
| 24 | SYK | 1212 | 5.837 | 0.2691 | Yes | ||
| 25 | TNFRSF4 | 1226 | 5.806 | 0.2786 | Yes | ||
| 26 | AKT2 | 1306 | 5.599 | 0.2848 | Yes | ||
| 27 | MCC | 1329 | 5.552 | 0.2935 | Yes | ||
| 28 | SH3BP2 | 1415 | 5.373 | 0.2990 | Yes | ||
| 29 | RGS16 | 1461 | 5.287 | 0.3061 | Yes | ||
| 30 | ITGB2 | 1471 | 5.262 | 0.3149 | Yes | ||
| 31 | AXIN1 | 1475 | 5.258 | 0.3240 | Yes | ||
| 32 | CAMK4 | 1688 | 4.913 | 0.3228 | Yes | ||
| 33 | STAT5A | 1759 | 4.813 | 0.3280 | Yes | ||
| 34 | RELB | 1827 | 4.711 | 0.3332 | Yes | ||
| 35 | MAP3K14 | 1956 | 4.529 | 0.3352 | Yes | ||
| 36 | ILK | 2044 | 4.405 | 0.3389 | Yes | ||
| 37 | ITGB4 | 2143 | 4.287 | 0.3419 | Yes | ||
| 38 | PDPK1 | 2170 | 4.258 | 0.3482 | Yes | ||
| 39 | MAP2K6 | 2378 | 4.033 | 0.3457 | Yes | ||
| 40 | ROCK1 | 2484 | 3.908 | 0.3477 | Yes | ||
| 41 | TNFRSF1B | 2492 | 3.901 | 0.3542 | Yes | ||
| 42 | TNFRSF14 | 2551 | 3.851 | 0.3583 | Yes | ||
| 43 | AKAP13 | 2553 | 3.851 | 0.3650 | Yes | ||
| 44 | SHC1 | 2716 | 3.694 | 0.3640 | Yes | ||
| 45 | BCL10 | 2773 | 3.637 | 0.3678 | Yes | ||
| 46 | TIAM1 | 2834 | 3.585 | 0.3713 | Yes | ||
| 47 | ATM | 2892 | 3.523 | 0.3749 | Yes | ||
| 48 | MAPK7 | 2940 | 3.482 | 0.3788 | Yes | ||
| 49 | MAPKAPK5 | 2957 | 3.470 | 0.3841 | Yes | ||
| 50 | ITGA6 | 3003 | 3.432 | 0.3881 | Yes | ||
| 51 | IL6ST | 3028 | 3.412 | 0.3929 | Yes | ||
| 52 | JAK3 | 3047 | 3.397 | 0.3980 | Yes | ||
| 53 | ITGB3 | 3098 | 3.349 | 0.4016 | Yes | ||
| 54 | ZAP70 | 3141 | 3.317 | 0.4055 | Yes | ||
| 55 | STAT6 | 3153 | 3.303 | 0.4107 | Yes | ||
| 56 | MAP3K7IP1 | 3197 | 3.262 | 0.4145 | Yes | ||
| 57 | ARHGAP1 | 3297 | 3.177 | 0.4155 | Yes | ||
| 58 | IRS2 | 3324 | 3.163 | 0.4198 | Yes | ||
| 59 | SOS1 | 3360 | 3.127 | 0.4237 | Yes | ||
| 60 | MAP3K4 | 3381 | 3.115 | 0.4282 | Yes | ||
| 61 | DLG1 | 3405 | 3.099 | 0.4326 | Yes | ||
| 62 | TANK | 3424 | 3.086 | 0.4371 | Yes | ||
| 63 | BRAF | 3459 | 3.052 | 0.4409 | Yes | ||
| 64 | SKIL | 3528 | 2.998 | 0.4430 | Yes | ||
| 65 | DGKA | 3551 | 2.979 | 0.4472 | Yes | ||
| 66 | IL18R1 | 3579 | 2.962 | 0.4511 | Yes | ||
| 67 | ITGAE | 3600 | 2.948 | 0.4554 | Yes | ||
| 68 | ITGAV | 3775 | 2.817 | 0.4523 | Yes | ||
| 69 | AKT1 | 3802 | 2.803 | 0.4560 | Yes | ||
| 70 | STAT1 | 3835 | 2.777 | 0.4594 | Yes | ||
| 71 | IFNGR1 | 3964 | 2.667 | 0.4582 | Yes | ||
| 72 | APC | 4019 | 2.635 | 0.4603 | Yes | ||
| 73 | JAG1 | 4046 | 2.617 | 0.4637 | Yes | ||
| 74 | IL12RB1 | 4056 | 2.611 | 0.4679 | Yes | ||
| 75 | CRKL | 4137 | 2.560 | 0.4687 | Yes | ||
| 76 | NCK1 | 4581 | 2.272 | 0.4523 | No | ||
| 77 | BIRC2 | 4594 | 2.264 | 0.4557 | No | ||
| 78 | ITGA3 | 4630 | 2.239 | 0.4580 | No | ||
| 79 | SRC | 4632 | 2.239 | 0.4619 | No | ||
| 80 | PIK3CG | 4859 | 2.117 | 0.4552 | No | ||
| 81 | AXL | 4918 | 2.088 | 0.4562 | No | ||
| 82 | RALBP1 | 4938 | 2.076 | 0.4589 | No | ||
| 83 | TTK | 5350 | 1.853 | 0.4433 | No | ||
| 84 | MAPK11 | 5462 | 1.797 | 0.4413 | No | ||
| 85 | MPL | 5640 | 1.705 | 0.4362 | No | ||
| 86 | JAK1 | 5653 | 1.700 | 0.4386 | No | ||
| 87 | PIK3C3 | 5706 | 1.681 | 0.4392 | No | ||
| 88 | MAP2K4 | 5823 | 1.626 | 0.4367 | No | ||
| 89 | TNFSF13 | 6014 | 1.536 | 0.4306 | No | ||
| 90 | TYK2 | 6101 | 1.504 | 0.4293 | No | ||
| 91 | NRAS | 6347 | 1.384 | 0.4205 | No | ||
| 92 | WNT2 | 6534 | 1.291 | 0.4142 | No | ||
| 93 | FYN | 6571 | 1.275 | 0.4148 | No | ||
| 94 | TRAF6 | 6611 | 1.253 | 0.4152 | No | ||
| 95 | AXIN2 | 6861 | 1.146 | 0.4058 | No | ||
| 96 | NF1 | 6995 | 1.084 | 0.4015 | No | ||
| 97 | GRB7 | 7077 | 1.049 | 0.3997 | No | ||
| 98 | ABL2 | 7112 | 1.032 | 0.3999 | No | ||
| 99 | MAP2K5 | 7135 | 1.023 | 0.4007 | No | ||
| 100 | MAP2K3 | 7606 | 0.826 | 0.3805 | No | ||
| 101 | ITGAX | 7644 | 0.809 | 0.3803 | No | ||
| 102 | IL15 | 8051 | 0.642 | 0.3627 | No | ||
| 103 | ROS1 | 8397 | 0.498 | 0.3478 | No | ||
| 104 | ITGB7 | 8521 | 0.444 | 0.3429 | No | ||
| 105 | IGFBP1 | 8707 | 0.363 | 0.3350 | No | ||
| 106 | WNT4 | 8968 | 0.248 | 0.3235 | No | ||
| 107 | NOTCH3 | 9024 | 0.222 | 0.3214 | No | ||
| 108 | REL | 9035 | 0.218 | 0.3213 | No | ||
| 109 | MUSK | 9181 | 0.160 | 0.3149 | No | ||
| 110 | ITGB8 | 9268 | 0.123 | 0.3112 | No | ||
| 111 | MAP3K8 | 10010 | -0.225 | 0.2776 | No | ||
| 112 | MYD88 | 10348 | -0.356 | 0.2627 | No | ||
| 113 | ITGAM | 10658 | -0.458 | 0.2493 | No | ||
| 114 | MKNK1 | 11007 | -0.582 | 0.2343 | No | ||
| 115 | INHA | 11336 | -0.692 | 0.2205 | No | ||
| 116 | ARHGEF16 | 11411 | -0.717 | 0.2183 | No | ||
| 117 | MAPK4 | 11540 | -0.763 | 0.2138 | No | ||
| 118 | CTNNB1 | 11600 | -0.784 | 0.2125 | No | ||
| 119 | BTK | 12025 | -0.929 | 0.1946 | No | ||
| 120 | TNFSF4 | 12257 | -0.993 | 0.1857 | No | ||
| 121 | ERG | 12732 | -1.135 | 0.1660 | No | ||
| 122 | BCR | 12777 | -1.150 | 0.1660 | No | ||
| 123 | IRS1 | 13166 | -1.265 | 0.1503 | No | ||
| 124 | ITGA2 | 13254 | -1.289 | 0.1486 | No | ||
| 125 | MAPKAPK2 | 13256 | -1.290 | 0.1508 | No | ||
| 126 | MAP2K1 | 13318 | -1.307 | 0.1503 | No | ||
| 127 | MAPK12 | 13362 | -1.319 | 0.1506 | No | ||
| 128 | IL1B | 13416 | -1.337 | 0.1505 | No | ||
| 129 | TNF | 13449 | -1.346 | 0.1514 | No | ||
| 130 | CXCL2 | 13573 | -1.387 | 0.1482 | No | ||
| 131 | JAG2 | 13706 | -1.429 | 0.1446 | No | ||
| 132 | TYRO3 | 14002 | -1.507 | 0.1337 | No | ||
| 133 | MAPK13 | 14111 | -1.538 | 0.1314 | No | ||
| 134 | MAPK9 | 14521 | -1.654 | 0.1156 | No | ||
| 135 | CXCL12 | 14528 | -1.656 | 0.1182 | No | ||
| 136 | ARHGEF12 | 14701 | -1.713 | 0.1133 | No | ||
| 137 | IGFBP3 | 14705 | -1.713 | 0.1161 | No | ||
| 138 | IL10 | 14877 | -1.762 | 0.1114 | No | ||
| 139 | ARHGAP6 | 14933 | -1.779 | 0.1119 | No | ||
| 140 | NOTCH4 | 14957 | -1.787 | 0.1140 | No | ||
| 141 | IL2RB | 15643 | -2.012 | 0.0861 | No | ||
| 142 | ITGA5 | 16195 | -2.180 | 0.0646 | No | ||
| 143 | ITGA1 | 16200 | -2.183 | 0.0682 | No | ||
| 144 | IFNG | 16549 | -2.307 | 0.0563 | No | ||
| 145 | RAP1A | 16633 | -2.333 | 0.0565 | No | ||
| 146 | VIPR1 | 16776 | -2.385 | 0.0542 | No | ||
| 147 | RASA1 | 16898 | -2.430 | 0.0529 | No | ||
| 148 | ITGA7 | 16908 | -2.433 | 0.0567 | No | ||
| 149 | WNT5A | 17341 | -2.593 | 0.0414 | No | ||
| 150 | IFNB1 | 17366 | -2.603 | 0.0449 | No | ||
| 151 | IL13 | 17963 | -2.838 | 0.0224 | No | ||
| 152 | SFN | 18116 | -2.905 | 0.0205 | No | ||
| 153 | HRAS | 18182 | -2.929 | 0.0227 | No | ||
| 154 | MAS1 | 18323 | -2.988 | 0.0215 | No | ||
| 155 | GNB1 | 18351 | -3.002 | 0.0255 | No | ||
| 156 | ITGB5 | 18507 | -3.077 | 0.0237 | No | ||
| 157 | SUFU | 18528 | -3.087 | 0.0282 | No | ||
| 158 | CALM1 | 18993 | -3.349 | 0.0128 | No | ||
| 159 | ITGA4 | 19179 | -3.470 | 0.0103 | No | ||
| 160 | GRB14 | 19214 | -3.487 | 0.0149 | No | ||
| 161 | PTCH2 | 19247 | -3.508 | 0.0195 | No | ||
| 162 | MAPK10 | 19290 | -3.532 | 0.0238 | No | ||
| 163 | WNT1 | 19449 | -3.646 | 0.0229 | No | ||
| 164 | CXCL1 | 19585 | -3.754 | 0.0233 | No | ||
| 165 | MAP3K12 | 19825 | -3.968 | 0.0192 | No | ||
| 166 | TNFRSF10B | 20131 | -4.274 | 0.0127 | No | ||
| 167 | SMO | 20258 | -4.408 | 0.0146 | No | ||
| 168 | MAPK1 | 20477 | -4.696 | 0.0128 | No | ||
| 169 | PTPRK | 20543 | -4.769 | 0.0182 | No | ||
| 170 | RAN | 21154 | -5.842 | 0.0004 | No | ||
| 171 | ITGB1 | 21199 | -5.975 | 0.0088 | No | ||
| 172 | CRK | 21299 | -6.288 | 0.0152 | No | ||
| 173 | ITGA9 | 21728 | -8.273 | 0.0101 | No |