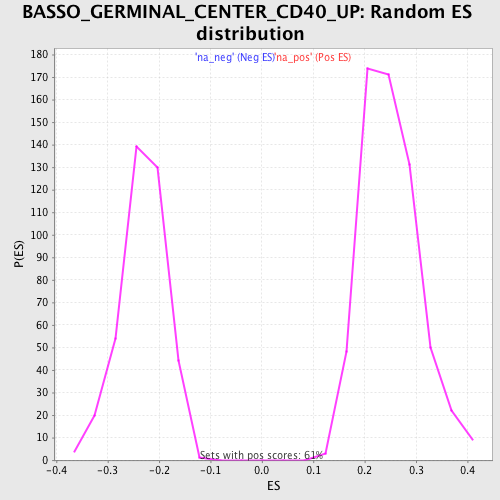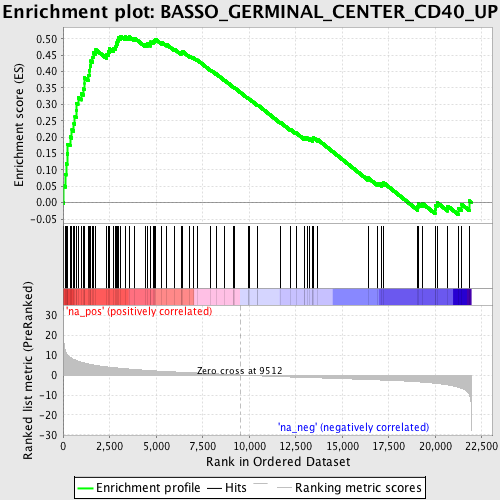
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | BASSO_GERMINAL_CENTER_CD40_UP |
| Enrichment Score (ES) | 0.5082637 |
| Normalized Enrichment Score (NES) | 2.0365043 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0101571465 |
| FWER p-Value | 0.116 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCR7 | 34 | 17.063 | 0.0514 | Yes | ||
| 2 | NFKBIA | 116 | 12.486 | 0.0865 | Yes | ||
| 3 | PTK2B | 159 | 11.293 | 0.1196 | Yes | ||
| 4 | PIK3CD | 211 | 10.436 | 0.1497 | Yes | ||
| 5 | ISGF3G | 251 | 9.973 | 0.1789 | Yes | ||
| 6 | JUNB | 376 | 8.916 | 0.2009 | Yes | ||
| 7 | TNFAIP3 | 474 | 8.304 | 0.2222 | Yes | ||
| 8 | MTMR4 | 579 | 7.763 | 0.2416 | Yes | ||
| 9 | LAPTM5 | 634 | 7.551 | 0.2625 | Yes | ||
| 10 | GADD45B | 712 | 7.204 | 0.2814 | Yes | ||
| 11 | ZFP36L1 | 720 | 7.180 | 0.3033 | Yes | ||
| 12 | TRAF1 | 801 | 6.898 | 0.3211 | Yes | ||
| 13 | DUSP2 | 971 | 6.410 | 0.3333 | Yes | ||
| 14 | CFLAR | 1080 | 6.128 | 0.3473 | Yes | ||
| 15 | BCL2 | 1151 | 5.973 | 0.3627 | Yes | ||
| 16 | TNFSF9 | 1154 | 5.967 | 0.3811 | Yes | ||
| 17 | DTX2 | 1354 | 5.496 | 0.3891 | Yes | ||
| 18 | NCKAP1L | 1404 | 5.391 | 0.4036 | Yes | ||
| 19 | NFKB2 | 1444 | 5.315 | 0.4183 | Yes | ||
| 20 | EBI2 | 1489 | 5.233 | 0.4325 | Yes | ||
| 21 | TCEB3 | 1593 | 5.064 | 0.4435 | Yes | ||
| 22 | KCNN4 | 1620 | 5.016 | 0.4579 | Yes | ||
| 23 | STAT5A | 1759 | 4.813 | 0.4665 | Yes | ||
| 24 | SLAMF1 | 2349 | 4.066 | 0.4522 | Yes | ||
| 25 | PLXNC1 | 2431 | 3.971 | 0.4608 | Yes | ||
| 26 | TMC6 | 2504 | 3.888 | 0.4696 | Yes | ||
| 27 | SNN | 2713 | 3.697 | 0.4716 | Yes | ||
| 28 | DDIT4 | 2835 | 3.583 | 0.4772 | Yes | ||
| 29 | CHD1 | 2853 | 3.562 | 0.4875 | Yes | ||
| 30 | STK10 | 2936 | 3.486 | 0.4945 | Yes | ||
| 31 | TAP1 | 2968 | 3.462 | 0.5039 | Yes | ||
| 32 | ARID5B | 3100 | 3.349 | 0.5083 | Yes | ||
| 33 | ICAM1 | 3353 | 3.133 | 0.5065 | No | ||
| 34 | ARHGEF2 | 3552 | 2.978 | 0.5067 | No | ||
| 35 | STAT1 | 3835 | 2.777 | 0.5024 | No | ||
| 36 | SNF1LK | 4417 | 2.374 | 0.4832 | No | ||
| 37 | BATF | 4535 | 2.304 | 0.4850 | No | ||
| 38 | CCL22 | 4689 | 2.205 | 0.4848 | No | ||
| 39 | PHACTR1 | 4715 | 2.193 | 0.4905 | No | ||
| 40 | LTA | 4836 | 2.132 | 0.4916 | No | ||
| 41 | RAC2 | 4891 | 2.101 | 0.4957 | No | ||
| 42 | RXRA | 4957 | 2.065 | 0.4991 | No | ||
| 43 | NFKBIE | 5291 | 1.879 | 0.4897 | No | ||
| 44 | EHD1 | 5548 | 1.750 | 0.4834 | No | ||
| 45 | MYB | 5989 | 1.550 | 0.4681 | No | ||
| 46 | PTP4A3 | 6340 | 1.387 | 0.4564 | No | ||
| 47 | CD83 | 6369 | 1.376 | 0.4594 | No | ||
| 48 | ELL2 | 6417 | 1.354 | 0.4614 | No | ||
| 49 | TMSB4X | 6803 | 1.172 | 0.4475 | No | ||
| 50 | CAPG | 6992 | 1.084 | 0.4422 | No | ||
| 51 | TRIP10 | 7208 | 0.995 | 0.4355 | No | ||
| 52 | CD40 | 7927 | 0.694 | 0.4048 | No | ||
| 53 | SH2B3 | 8222 | 0.568 | 0.3931 | No | ||
| 54 | HTR3A | 8697 | 0.369 | 0.3726 | No | ||
| 55 | CR2 | 9161 | 0.166 | 0.3519 | No | ||
| 56 | LGALS1 | 9176 | 0.161 | 0.3518 | No | ||
| 57 | CCL4 | 9220 | 0.140 | 0.3502 | No | ||
| 58 | NCF2 | 9952 | -0.199 | 0.3174 | No | ||
| 59 | CCL3 | 9968 | -0.204 | 0.3174 | No | ||
| 60 | MAP3K8 | 10010 | -0.225 | 0.3162 | No | ||
| 61 | RGS1 | 10450 | -0.389 | 0.2973 | No | ||
| 62 | HSP90B1 | 10470 | -0.395 | 0.2977 | No | ||
| 63 | FAIM3 | 11689 | -0.821 | 0.2445 | No | ||
| 64 | ADAM8 | 12242 | -0.989 | 0.2223 | No | ||
| 65 | IRF5 | 12519 | -1.072 | 0.2130 | No | ||
| 66 | IRF4 | 12943 | -1.197 | 0.1974 | No | ||
| 67 | TCFL5 | 12974 | -1.208 | 0.1998 | No | ||
| 68 | LITAF | 13107 | -1.247 | 0.1976 | No | ||
| 69 | MAPKAPK2 | 13256 | -1.290 | 0.1948 | No | ||
| 70 | IL1B | 13416 | -1.337 | 0.1917 | No | ||
| 71 | NEF3 | 13424 | -1.340 | 0.1956 | No | ||
| 72 | TNF | 13449 | -1.346 | 0.1986 | No | ||
| 73 | BLR1 | 13686 | -1.423 | 0.1923 | No | ||
| 74 | IFI30 | 16382 | -2.243 | 0.0759 | No | ||
| 75 | ICOSLG | 16894 | -2.429 | 0.0601 | No | ||
| 76 | CD44 | 17112 | -2.503 | 0.0580 | No | ||
| 77 | MEF2C | 17220 | -2.542 | 0.0610 | No | ||
| 78 | CD74 | 19057 | -3.386 | -0.0125 | No | ||
| 79 | TNFAIP8 | 19073 | -3.400 | -0.0027 | No | ||
| 80 | IER3 | 19295 | -3.536 | -0.0018 | No | ||
| 81 | LHFPL2 | 19988 | -4.123 | -0.0206 | No | ||
| 82 | RAB27A | 20022 | -4.167 | -0.0092 | No | ||
| 83 | PLEK | 20096 | -4.232 | 0.0006 | No | ||
| 84 | PRG1 | 20666 | -4.942 | -0.0101 | No | ||
| 85 | SYNGR2 | 21231 | -6.083 | -0.0170 | No | ||
| 86 | TNFAIP2 | 21397 | -6.563 | -0.0042 | No | ||
| 87 | UNC119 | 21822 | -9.436 | 0.0057 | No |