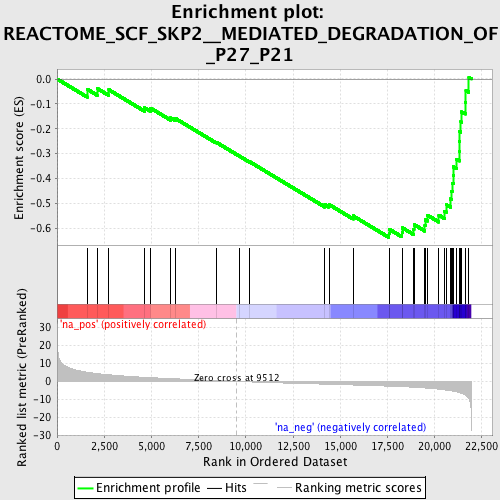
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_SCF_SKP2__MEDIATED_DEGRADATION_OF_P27_P21 |
| Enrichment Score (ES) | -0.63672304 |
| Normalized Enrichment Score (NES) | -2.3545353 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMB4 | 1590 | 5.066 | -0.0408 | No | ||
| 2 | CDK2 | 2120 | 4.313 | -0.0379 | No | ||
| 3 | PSMC4 | 2744 | 3.664 | -0.0433 | No | ||
| 4 | PSMD4 | 4636 | 2.236 | -0.1156 | No | ||
| 5 | CUL1 | 4951 | 2.070 | -0.1169 | No | ||
| 6 | PSMD10 | 6026 | 1.530 | -0.1564 | No | ||
| 7 | PSMB6 | 6284 | 1.412 | -0.1592 | No | ||
| 8 | PSMB5 | 8431 | 0.485 | -0.2541 | No | ||
| 9 | PSMD2 | 9671 | -0.075 | -0.3102 | No | ||
| 10 | PSMB3 | 10201 | -0.297 | -0.3325 | No | ||
| 11 | PSMD8 | 14190 | -1.564 | -0.5047 | No | ||
| 12 | PSMC3 | 14407 | -1.622 | -0.5044 | No | ||
| 13 | PSMD3 | 15684 | -2.029 | -0.5499 | No | ||
| 14 | PSMB9 | 17587 | -2.677 | -0.6199 | Yes | ||
| 15 | PSMA5 | 17619 | -2.689 | -0.6045 | Yes | ||
| 16 | CDKN1A | 18276 | -2.964 | -0.6158 | Yes | ||
| 17 | PSMD9 | 18302 | -2.978 | -0.5983 | Yes | ||
| 18 | PSMA3 | 18882 | -3.284 | -0.6041 | Yes | ||
| 19 | PSMA4 | 18913 | -3.305 | -0.5847 | Yes | ||
| 20 | PSMC1 | 19490 | -3.677 | -0.5879 | Yes | ||
| 21 | PSMD6 | 19506 | -3.694 | -0.5655 | Yes | ||
| 22 | PSMC5 | 19623 | -3.785 | -0.5470 | Yes | ||
| 23 | PSME1 | 20232 | -4.389 | -0.5472 | Yes | ||
| 24 | PSMA6 | 20539 | -4.766 | -0.5313 | Yes | ||
| 25 | PSMB10 | 20617 | -4.853 | -0.5043 | Yes | ||
| 26 | PSMA7 | 20850 | -5.266 | -0.4819 | Yes | ||
| 27 | PSMD12 | 20898 | -5.320 | -0.4507 | Yes | ||
| 28 | PSMD5 | 20934 | -5.370 | -0.4186 | Yes | ||
| 29 | PSMB1 | 20998 | -5.482 | -0.3870 | Yes | ||
| 30 | PSMB2 | 21012 | -5.515 | -0.3530 | Yes | ||
| 31 | PSMD14 | 21175 | -5.898 | -0.3234 | Yes | ||
| 32 | PSMA1 | 21311 | -6.328 | -0.2899 | Yes | ||
| 33 | PSMD11 | 21321 | -6.341 | -0.2505 | Yes | ||
| 34 | PSMD7 | 21338 | -6.407 | -0.2110 | Yes | ||
| 35 | PSMA2 | 21396 | -6.560 | -0.1725 | Yes | ||
| 36 | CKS1B | 21442 | -6.729 | -0.1323 | Yes | ||
| 37 | PSMC2 | 21627 | -7.542 | -0.0934 | Yes | ||
| 38 | PSMC6 | 21665 | -7.755 | -0.0464 | Yes | ||
| 39 | PSME3 | 21823 | -9.439 | 0.0057 | Yes |