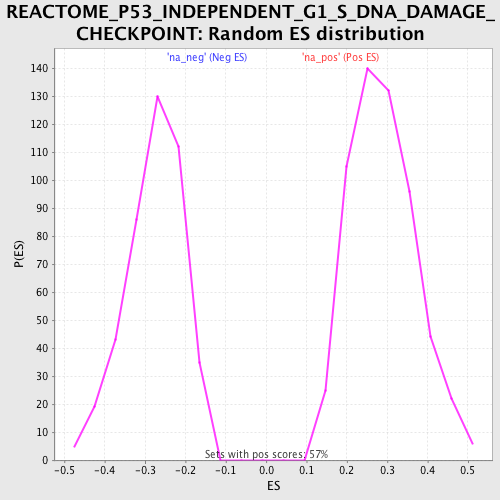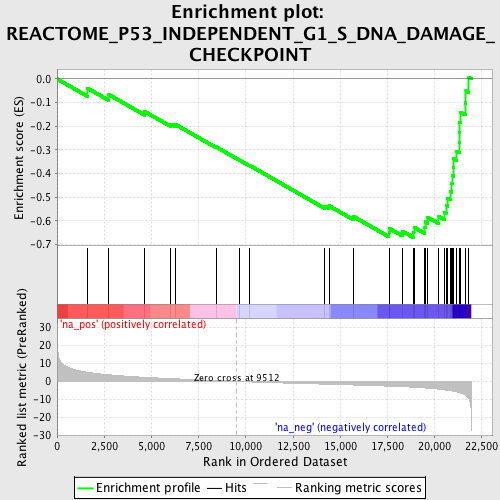
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_P53_INDEPENDENT_G1_S_DNA_DAMAGE_CHECKPOINT |
| Enrichment Score (ES) | -0.6700714 |
| Normalized Enrichment Score (NES) | -2.4195802 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMB4 | 1590 | 5.066 | -0.0384 | No | ||
| 2 | PSMC4 | 2744 | 3.664 | -0.0663 | No | ||
| 3 | PSMD4 | 4636 | 2.236 | -0.1375 | No | ||
| 4 | PSMD10 | 6026 | 1.530 | -0.1906 | No | ||
| 5 | PSMB6 | 6284 | 1.412 | -0.1928 | No | ||
| 6 | PSMB5 | 8431 | 0.485 | -0.2875 | No | ||
| 7 | PSMD2 | 9671 | -0.075 | -0.3435 | No | ||
| 8 | PSMB3 | 10201 | -0.297 | -0.3657 | No | ||
| 9 | PSMD8 | 14190 | -1.564 | -0.5371 | No | ||
| 10 | PSMC3 | 14407 | -1.622 | -0.5360 | No | ||
| 11 | PSMD3 | 15684 | -2.029 | -0.5806 | No | ||
| 12 | PSMB9 | 17587 | -2.677 | -0.6493 | Yes | ||
| 13 | PSMA5 | 17619 | -2.689 | -0.6326 | Yes | ||
| 14 | PSMD9 | 18302 | -2.978 | -0.6436 | Yes | ||
| 15 | PSMA3 | 18882 | -3.284 | -0.6479 | Yes | ||
| 16 | PSMA4 | 18913 | -3.305 | -0.6270 | Yes | ||
| 17 | PSMC1 | 19490 | -3.677 | -0.6285 | Yes | ||
| 18 | PSMD6 | 19506 | -3.694 | -0.6043 | Yes | ||
| 19 | PSMC5 | 19623 | -3.785 | -0.5840 | Yes | ||
| 20 | PSME1 | 20232 | -4.389 | -0.5822 | Yes | ||
| 21 | PSMA6 | 20539 | -4.766 | -0.5640 | Yes | ||
| 22 | PSMB10 | 20617 | -4.853 | -0.5348 | Yes | ||
| 23 | CDC25A | 20686 | -4.987 | -0.5043 | Yes | ||
| 24 | PSMA7 | 20850 | -5.266 | -0.4762 | Yes | ||
| 25 | PSMD12 | 20898 | -5.320 | -0.4425 | Yes | ||
| 26 | PSMD5 | 20934 | -5.370 | -0.4079 | Yes | ||
| 27 | PSMB1 | 20998 | -5.482 | -0.3738 | Yes | ||
| 28 | PSMB2 | 21012 | -5.515 | -0.3372 | Yes | ||
| 29 | PSMD14 | 21175 | -5.898 | -0.3048 | Yes | ||
| 30 | PSMA1 | 21311 | -6.328 | -0.2683 | Yes | ||
| 31 | PSMD11 | 21321 | -6.341 | -0.2259 | Yes | ||
| 32 | PSMD7 | 21338 | -6.407 | -0.1834 | Yes | ||
| 33 | PSMA2 | 21396 | -6.560 | -0.1418 | Yes | ||
| 34 | PSMC2 | 21627 | -7.542 | -0.1014 | Yes | ||
| 35 | PSMC6 | 21665 | -7.755 | -0.0508 | Yes | ||
| 36 | PSME3 | 21823 | -9.439 | 0.0057 | Yes |