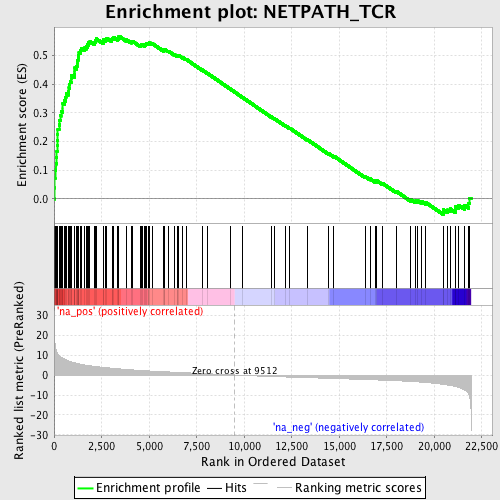
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | NETPATH_TCR |
| Enrichment Score (ES) | 0.56730807 |
| Normalized Enrichment Score (NES) | 2.418373 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PRKD2 | 9 | 20.994 | 0.0384 | Yes | ||
| 2 | LCK | 20 | 19.234 | 0.0736 | Yes | ||
| 3 | CD3E | 51 | 15.588 | 0.1011 | Yes | ||
| 4 | SIT1 | 99 | 12.922 | 0.1228 | Yes | ||
| 5 | STAT5B | 125 | 12.304 | 0.1445 | Yes | ||
| 6 | GRAP | 146 | 11.545 | 0.1649 | Yes | ||
| 7 | PTK2B | 159 | 11.293 | 0.1853 | Yes | ||
| 8 | WASF2 | 168 | 11.019 | 0.2053 | Yes | ||
| 9 | GIT2 | 191 | 10.702 | 0.2241 | Yes | ||
| 10 | CD247 | 200 | 10.587 | 0.2433 | Yes | ||
| 11 | CD8A | 263 | 9.816 | 0.2586 | Yes | ||
| 12 | CD4 | 297 | 9.537 | 0.2748 | Yes | ||
| 13 | SKAP1 | 331 | 9.264 | 0.2904 | Yes | ||
| 14 | ARHGDIB | 365 | 8.988 | 0.3055 | Yes | ||
| 15 | DNM2 | 460 | 8.378 | 0.3167 | Yes | ||
| 16 | ABL1 | 461 | 8.376 | 0.3322 | Yes | ||
| 17 | PLCG1 | 539 | 7.963 | 0.3435 | Yes | ||
| 18 | CISH | 606 | 7.660 | 0.3546 | Yes | ||
| 19 | VAV1 | 628 | 7.582 | 0.3677 | Yes | ||
| 20 | EVL | 753 | 7.045 | 0.3750 | Yes | ||
| 21 | DEF6 | 781 | 6.943 | 0.3867 | Yes | ||
| 22 | PSTPIP1 | 804 | 6.893 | 0.3984 | Yes | ||
| 23 | CARD11 | 836 | 6.773 | 0.4095 | Yes | ||
| 24 | LAX1 | 898 | 6.608 | 0.4190 | Yes | ||
| 25 | SH2D3C | 905 | 6.573 | 0.4309 | Yes | ||
| 26 | CD3D | 1059 | 6.186 | 0.4353 | Yes | ||
| 27 | ITK | 1091 | 6.101 | 0.4452 | Yes | ||
| 28 | TXK | 1094 | 6.095 | 0.4564 | Yes | ||
| 29 | VASP | 1190 | 5.873 | 0.4629 | Yes | ||
| 30 | SYK | 1212 | 5.837 | 0.4727 | Yes | ||
| 31 | CBL | 1220 | 5.822 | 0.4832 | Yes | ||
| 32 | DOCK2 | 1262 | 5.718 | 0.4919 | Yes | ||
| 33 | CREB1 | 1283 | 5.654 | 0.5014 | Yes | ||
| 34 | SLA2 | 1307 | 5.599 | 0.5107 | Yes | ||
| 35 | LCP2 | 1403 | 5.391 | 0.5164 | Yes | ||
| 36 | SH3BP2 | 1415 | 5.373 | 0.5258 | Yes | ||
| 37 | GAB2 | 1612 | 5.026 | 0.5261 | Yes | ||
| 38 | PAG1 | 1698 | 4.900 | 0.5313 | Yes | ||
| 39 | STAT5A | 1759 | 4.813 | 0.5375 | Yes | ||
| 40 | RAPGEF1 | 1794 | 4.763 | 0.5447 | Yes | ||
| 41 | CBLB | 1885 | 4.615 | 0.5492 | Yes | ||
| 42 | TUBB2A | 2136 | 4.298 | 0.5457 | Yes | ||
| 43 | CD5 | 2179 | 4.244 | 0.5516 | Yes | ||
| 44 | CREBBP | 2209 | 4.217 | 0.5581 | Yes | ||
| 45 | SLA | 2586 | 3.823 | 0.5479 | Yes | ||
| 46 | JUN | 2591 | 3.817 | 0.5548 | Yes | ||
| 47 | SHC1 | 2716 | 3.694 | 0.5560 | Yes | ||
| 48 | BCL10 | 2773 | 3.637 | 0.5601 | Yes | ||
| 49 | PTPRC | 3046 | 3.398 | 0.5540 | Yes | ||
| 50 | JAK3 | 3047 | 3.397 | 0.5603 | Yes | ||
| 51 | ZAP70 | 3141 | 3.317 | 0.5621 | Yes | ||
| 52 | SOS1 | 3360 | 3.127 | 0.5579 | Yes | ||
| 53 | DLG1 | 3405 | 3.099 | 0.5617 | Yes | ||
| 54 | NEDD9 | 3408 | 3.097 | 0.5673 | Yes | ||
| 55 | AKT1 | 3802 | 2.803 | 0.5545 | No | ||
| 56 | CD3G | 4051 | 2.614 | 0.5480 | No | ||
| 57 | CRKL | 4137 | 2.560 | 0.5488 | No | ||
| 58 | PTPN6 | 4545 | 2.295 | 0.5344 | No | ||
| 59 | NCK1 | 4581 | 2.272 | 0.5370 | No | ||
| 60 | SRC | 4632 | 2.239 | 0.5389 | No | ||
| 61 | TRAT1 | 4780 | 2.154 | 0.5362 | No | ||
| 62 | HOMER3 | 4830 | 2.135 | 0.5379 | No | ||
| 63 | ARHGEF7 | 4847 | 2.125 | 0.5411 | No | ||
| 64 | DBNL | 4946 | 2.073 | 0.5404 | No | ||
| 65 | SH2D2A | 4976 | 2.050 | 0.5429 | No | ||
| 66 | PTK2 | 5034 | 2.014 | 0.5440 | No | ||
| 67 | PTPN12 | 5165 | 1.945 | 0.5416 | No | ||
| 68 | PIK3R1 | 5770 | 1.654 | 0.5170 | No | ||
| 69 | PTPN11 | 5785 | 1.643 | 0.5194 | No | ||
| 70 | GRAP2 | 5821 | 1.627 | 0.5209 | No | ||
| 71 | ARHGEF6 | 6013 | 1.536 | 0.5150 | No | ||
| 72 | LIME1 | 6312 | 1.399 | 0.5039 | No | ||
| 73 | PRKCQ | 6476 | 1.323 | 0.4989 | No | ||
| 74 | ABI1 | 6483 | 1.320 | 0.5010 | No | ||
| 75 | FYN | 6571 | 1.275 | 0.4994 | No | ||
| 76 | FOS | 6777 | 1.183 | 0.4922 | No | ||
| 77 | WAS | 6943 | 1.105 | 0.4867 | No | ||
| 78 | SOS2 | 7785 | 0.747 | 0.4496 | No | ||
| 79 | KHDRBS1 | 8076 | 0.631 | 0.4375 | No | ||
| 80 | LAT | 9292 | 0.110 | 0.3820 | No | ||
| 81 | FYB | 9891 | -0.172 | 0.3550 | No | ||
| 82 | SKAP2 | 11417 | -0.720 | 0.2865 | No | ||
| 83 | CTNNB1 | 11600 | -0.784 | 0.2796 | No | ||
| 84 | PTPN3 | 12190 | -0.972 | 0.2544 | No | ||
| 85 | VAV3 | 12393 | -1.035 | 0.2471 | No | ||
| 86 | MAP2K1 | 13318 | -1.307 | 0.2072 | No | ||
| 87 | PTPN22 | 14441 | -1.630 | 0.1588 | No | ||
| 88 | NFAM1 | 14728 | -1.721 | 0.1489 | No | ||
| 89 | ACP1 | 16366 | -2.238 | 0.0781 | No | ||
| 90 | RAP1A | 16633 | -2.333 | 0.0703 | No | ||
| 91 | RASA1 | 16898 | -2.430 | 0.0627 | No | ||
| 92 | RIPK2 | 16962 | -2.453 | 0.0643 | No | ||
| 93 | CD2AP | 17306 | -2.577 | 0.0534 | No | ||
| 94 | STK39 | 18018 | -2.863 | 0.0261 | No | ||
| 95 | DUSP3 | 18761 | -3.218 | -0.0019 | No | ||
| 96 | PIK3R2 | 19008 | -3.358 | -0.0069 | No | ||
| 97 | CDC42 | 19144 | -3.447 | -0.0067 | No | ||
| 98 | PTPRJ | 19346 | -3.571 | -0.0093 | No | ||
| 99 | NFATC2 | 19548 | -3.725 | -0.0116 | No | ||
| 100 | MAPK1 | 20477 | -4.696 | -0.0455 | No | ||
| 101 | ENAH | 20485 | -4.704 | -0.0371 | No | ||
| 102 | DTX1 | 20720 | -5.069 | -0.0384 | No | ||
| 103 | GRB2 | 20842 | -5.251 | -0.0342 | No | ||
| 104 | SHB | 21116 | -5.744 | -0.0361 | No | ||
| 105 | YWHAQ | 21137 | -5.795 | -0.0263 | No | ||
| 106 | CRK | 21299 | -6.288 | -0.0220 | No | ||
| 107 | NCL | 21602 | -7.341 | -0.0223 | No | ||
| 108 | UNC119 | 21822 | -9.436 | -0.0148 | No | ||
| 109 | CEBPB | 21876 | -11.074 | 0.0033 | No |