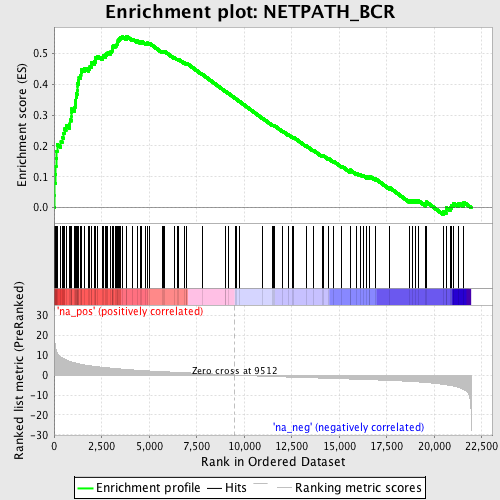
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | 0.5557317 |
| Normalized Enrichment Score (NES) | 2.3707561 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CASP7 | 14 | 20.405 | 0.0406 | Yes | ||
| 2 | LCK | 20 | 19.234 | 0.0792 | Yes | ||
| 3 | CCND3 | 59 | 14.931 | 0.1076 | Yes | ||
| 4 | BCL6 | 77 | 13.547 | 0.1342 | Yes | ||
| 5 | TEC | 104 | 12.753 | 0.1587 | Yes | ||
| 6 | NFKBIA | 116 | 12.486 | 0.1835 | Yes | ||
| 7 | PTK2B | 159 | 11.293 | 0.2043 | Yes | ||
| 8 | DAPP1 | 361 | 9.005 | 0.2133 | Yes | ||
| 9 | VAV2 | 421 | 8.556 | 0.2279 | Yes | ||
| 10 | ACTR2 | 500 | 8.150 | 0.2408 | Yes | ||
| 11 | PLCG1 | 539 | 7.963 | 0.2551 | Yes | ||
| 12 | VAV1 | 628 | 7.582 | 0.2664 | Yes | ||
| 13 | NFATC1 | 826 | 6.800 | 0.2711 | Yes | ||
| 14 | CARD11 | 836 | 6.773 | 0.2844 | Yes | ||
| 15 | RELA | 916 | 6.549 | 0.2940 | Yes | ||
| 16 | CCND2 | 921 | 6.537 | 0.3070 | Yes | ||
| 17 | ITPR2 | 927 | 6.521 | 0.3199 | Yes | ||
| 18 | ITK | 1091 | 6.101 | 0.3248 | Yes | ||
| 19 | IKBKG | 1106 | 6.072 | 0.3364 | Yes | ||
| 20 | GAB1 | 1132 | 6.018 | 0.3474 | Yes | ||
| 21 | BCL2 | 1151 | 5.973 | 0.3587 | Yes | ||
| 22 | NFATC3 | 1178 | 5.913 | 0.3694 | Yes | ||
| 23 | SYK | 1212 | 5.837 | 0.3797 | Yes | ||
| 24 | CBL | 1220 | 5.822 | 0.3911 | Yes | ||
| 25 | CSK | 1223 | 5.821 | 0.4028 | Yes | ||
| 26 | CREB1 | 1283 | 5.654 | 0.4115 | Yes | ||
| 27 | SLA2 | 1307 | 5.599 | 0.4217 | Yes | ||
| 28 | LCP2 | 1403 | 5.391 | 0.4283 | Yes | ||
| 29 | SH3BP2 | 1415 | 5.373 | 0.4386 | Yes | ||
| 30 | CD79B | 1433 | 5.349 | 0.4487 | Yes | ||
| 31 | GAB2 | 1612 | 5.026 | 0.4506 | Yes | ||
| 32 | RAPGEF1 | 1794 | 4.763 | 0.4520 | Yes | ||
| 33 | CBLB | 1885 | 4.615 | 0.4572 | Yes | ||
| 34 | RB1 | 1959 | 4.524 | 0.4630 | Yes | ||
| 35 | MAPK3 | 1966 | 4.511 | 0.4718 | Yes | ||
| 36 | CDK2 | 2120 | 4.313 | 0.4735 | Yes | ||
| 37 | PDPK1 | 2170 | 4.258 | 0.4799 | Yes | ||
| 38 | CD5 | 2179 | 4.244 | 0.4881 | Yes | ||
| 39 | HCLS1 | 2300 | 4.113 | 0.4909 | Yes | ||
| 40 | HDAC5 | 2567 | 3.840 | 0.4864 | Yes | ||
| 41 | JUN | 2591 | 3.817 | 0.4931 | Yes | ||
| 42 | SHC1 | 2716 | 3.694 | 0.4949 | Yes | ||
| 43 | BCL10 | 2773 | 3.637 | 0.4996 | Yes | ||
| 44 | CDK6 | 2833 | 3.585 | 0.5042 | Yes | ||
| 45 | GSK3B | 2980 | 3.451 | 0.5045 | Yes | ||
| 46 | PTPRC | 3046 | 3.398 | 0.5083 | Yes | ||
| 47 | ATF2 | 3068 | 3.373 | 0.5142 | Yes | ||
| 48 | MAPK14 | 3074 | 3.368 | 0.5208 | Yes | ||
| 49 | ZAP70 | 3141 | 3.317 | 0.5244 | Yes | ||
| 50 | PTPN18 | 3232 | 3.238 | 0.5269 | Yes | ||
| 51 | DOK1 | 3306 | 3.173 | 0.5299 | Yes | ||
| 52 | RPS6KB1 | 3354 | 3.133 | 0.5341 | Yes | ||
| 53 | SOS1 | 3360 | 3.127 | 0.5402 | Yes | ||
| 54 | NEDD9 | 3408 | 3.097 | 0.5443 | Yes | ||
| 55 | BRAF | 3459 | 3.052 | 0.5482 | Yes | ||
| 56 | CD81 | 3504 | 3.020 | 0.5522 | Yes | ||
| 57 | BCL2L11 | 3585 | 2.958 | 0.5545 | Yes | ||
| 58 | AKT1 | 3802 | 2.803 | 0.5503 | Yes | ||
| 59 | PDK2 | 3808 | 2.796 | 0.5557 | Yes | ||
| 60 | CRKL | 4137 | 2.560 | 0.5459 | No | ||
| 61 | BCAR1 | 4367 | 2.400 | 0.5402 | No | ||
| 62 | PTPN6 | 4545 | 2.295 | 0.5368 | No | ||
| 63 | NCK1 | 4581 | 2.272 | 0.5397 | No | ||
| 64 | PPP3R1 | 4827 | 2.135 | 0.5328 | No | ||
| 65 | ELK1 | 4900 | 2.097 | 0.5338 | No | ||
| 66 | PTK2 | 5034 | 2.014 | 0.5317 | No | ||
| 67 | GSK3A | 5701 | 1.683 | 0.5046 | No | ||
| 68 | PIK3R1 | 5770 | 1.654 | 0.5048 | No | ||
| 69 | PTPN11 | 5785 | 1.643 | 0.5075 | No | ||
| 70 | LIME1 | 6312 | 1.399 | 0.4863 | No | ||
| 71 | PRKCQ | 6476 | 1.323 | 0.4815 | No | ||
| 72 | FYN | 6571 | 1.275 | 0.4797 | No | ||
| 73 | PRKCD | 6849 | 1.149 | 0.4694 | No | ||
| 74 | WAS | 6943 | 1.105 | 0.4673 | No | ||
| 75 | CHUK | 6991 | 1.084 | 0.4674 | No | ||
| 76 | SOS2 | 7785 | 0.747 | 0.4325 | No | ||
| 77 | PLEKHA1 | 8993 | 0.235 | 0.3777 | No | ||
| 78 | REL | 9035 | 0.218 | 0.3763 | No | ||
| 79 | CR2 | 9161 | 0.166 | 0.3709 | No | ||
| 80 | PIP5K1C | 9535 | -0.007 | 0.3538 | No | ||
| 81 | DOK3 | 9621 | -0.055 | 0.3500 | No | ||
| 82 | LAT2 | 9762 | -0.112 | 0.3438 | No | ||
| 83 | CD19 | 10959 | -0.570 | 0.2902 | No | ||
| 84 | RPS6 | 11490 | -0.745 | 0.2674 | No | ||
| 85 | MAPK4 | 11540 | -0.763 | 0.2667 | No | ||
| 86 | CTNNB1 | 11600 | -0.784 | 0.2656 | No | ||
| 87 | BTK | 12025 | -0.929 | 0.2480 | No | ||
| 88 | CCNA2 | 12324 | -1.015 | 0.2364 | No | ||
| 89 | PRKD1 | 12547 | -1.079 | 0.2284 | No | ||
| 90 | CD22 | 12622 | -1.103 | 0.2273 | No | ||
| 91 | MAPKAPK2 | 13256 | -1.290 | 0.2009 | No | ||
| 92 | ATP2B4 | 13634 | -1.402 | 0.1865 | No | ||
| 93 | HCK | 14105 | -1.536 | 0.1680 | No | ||
| 94 | GTF2I | 14162 | -1.555 | 0.1686 | No | ||
| 95 | ACTR3 | 14464 | -1.637 | 0.1581 | No | ||
| 96 | PIK3AP1 | 14700 | -1.713 | 0.1508 | No | ||
| 97 | CD79A | 15127 | -1.844 | 0.1350 | No | ||
| 98 | RASGRP3 | 15577 | -1.992 | 0.1185 | No | ||
| 99 | RPS6KA1 | 15581 | -1.993 | 0.1223 | No | ||
| 100 | BLNK | 15915 | -2.099 | 0.1113 | No | ||
| 101 | PRKCE | 16103 | -2.152 | 0.1071 | No | ||
| 102 | CASP9 | 16271 | -2.209 | 0.1039 | No | ||
| 103 | RAP2A | 16464 | -2.278 | 0.0997 | No | ||
| 104 | CCNE1 | 16580 | -2.317 | 0.0991 | No | ||
| 105 | CDK7 | 16622 | -2.330 | 0.1020 | No | ||
| 106 | RASA1 | 16898 | -2.430 | 0.0943 | No | ||
| 107 | CD72 | 17655 | -2.708 | 0.0651 | No | ||
| 108 | PPP3CA | 18699 | -3.178 | 0.0237 | No | ||
| 109 | MAP3K7 | 18867 | -3.272 | 0.0227 | No | ||
| 110 | PIK3R2 | 19008 | -3.358 | 0.0231 | No | ||
| 111 | MAPK8 | 19156 | -3.453 | 0.0233 | No | ||
| 112 | NFATC2 | 19548 | -3.725 | 0.0129 | No | ||
| 113 | IKBKB | 19586 | -3.755 | 0.0188 | No | ||
| 114 | MAPK1 | 20477 | -4.696 | -0.0125 | No | ||
| 115 | BAX | 20631 | -4.885 | -0.0096 | No | ||
| 116 | CYCS | 20634 | -4.887 | 0.0001 | No | ||
| 117 | GRB2 | 20842 | -5.251 | 0.0013 | No | ||
| 118 | BLK | 20922 | -5.356 | 0.0085 | No | ||
| 119 | CDK4 | 21020 | -5.528 | 0.0152 | No | ||
| 120 | CRK | 21299 | -6.288 | 0.0151 | No | ||
| 121 | FCGR2B | 21564 | -7.183 | 0.0175 | No |