Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$YY1_02 |
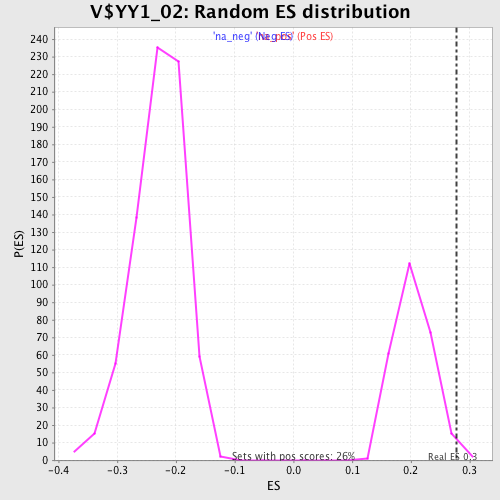
| Enrichment Score (ES) | 0.27752528 |
| Normalized Enrichment Score (NES) | 1.3616049 |
| Nominal p-value | 0.015151516 |
| FDR q-value | 0.21275881 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | UTX | 22 | 4.566 | 0.0217 | Yes | ||
| 2 | FOXO3A | 134 | 3.219 | 0.0327 | Yes | ||
| 3 | PPP1R12B | 224 | 2.836 | 0.0427 | Yes | ||
| 4 | ASH1L | 314 | 2.583 | 0.0515 | Yes | ||
| 5 | SNX13 | 354 | 2.510 | 0.0622 | Yes | ||
| 6 | TIA1 | 358 | 2.505 | 0.0745 | Yes | ||
| 7 | COL4A3BP | 445 | 2.359 | 0.0823 | Yes | ||
| 8 | ANAPC10 | 455 | 2.334 | 0.0935 | Yes | ||
| 9 | STRN3 | 477 | 2.310 | 0.1041 | Yes | ||
| 10 | UBE2J2 | 489 | 2.290 | 0.1150 | Yes | ||
| 11 | PPP2CA | 652 | 2.103 | 0.1180 | Yes | ||
| 12 | TNRC15 | 668 | 2.088 | 0.1277 | Yes | ||
| 13 | SFRS10 | 698 | 2.057 | 0.1366 | Yes | ||
| 14 | EPC1 | 736 | 2.022 | 0.1450 | Yes | ||
| 15 | POLK | 872 | 1.906 | 0.1483 | Yes | ||
| 16 | NCOR1 | 985 | 1.831 | 0.1523 | Yes | ||
| 17 | BCL11B | 1065 | 1.776 | 0.1575 | Yes | ||
| 18 | ASXL2 | 1084 | 1.767 | 0.1655 | Yes | ||
| 19 | ATE1 | 1146 | 1.734 | 0.1713 | Yes | ||
| 20 | PCF11 | 1156 | 1.729 | 0.1795 | Yes | ||
| 21 | PEG3 | 1302 | 1.652 | 0.1811 | Yes | ||
| 22 | DEPDC6 | 1313 | 1.648 | 0.1888 | Yes | ||
| 23 | EEF2 | 1317 | 1.647 | 0.1969 | Yes | ||
| 24 | TCF4 | 1328 | 1.641 | 0.2046 | Yes | ||
| 25 | TNKS2 | 1370 | 1.618 | 0.2108 | Yes | ||
| 26 | CHD4 | 1458 | 1.578 | 0.2146 | Yes | ||
| 27 | MLL5 | 1502 | 1.561 | 0.2204 | Yes | ||
| 28 | YWHAE | 1550 | 1.540 | 0.2260 | Yes | ||
| 29 | KLF7 | 1567 | 1.535 | 0.2329 | Yes | ||
| 30 | PTBP2 | 1577 | 1.531 | 0.2401 | Yes | ||
| 31 | RAB1A | 1738 | 1.468 | 0.2400 | Yes | ||
| 32 | EXT1 | 1759 | 1.458 | 0.2464 | Yes | ||
| 33 | MRPL1 | 1786 | 1.443 | 0.2524 | Yes | ||
| 34 | MTMR2 | 1914 | 1.387 | 0.2535 | Yes | ||
| 35 | DDX6 | 1948 | 1.376 | 0.2588 | Yes | ||
| 36 | ARCN1 | 2075 | 1.336 | 0.2597 | Yes | ||
| 37 | BCL2L1 | 2231 | 1.283 | 0.2589 | Yes | ||
| 38 | SLITRK5 | 2249 | 1.277 | 0.2645 | Yes | ||
| 39 | PHC3 | 2318 | 1.253 | 0.2676 | Yes | ||
| 40 | GTF2A1 | 2485 | 1.202 | 0.2660 | Yes | ||
| 41 | ZZEF1 | 2767 | 1.135 | 0.2588 | Yes | ||
| 42 | ZFX | 2804 | 1.126 | 0.2627 | Yes | ||
| 43 | POU2F1 | 2833 | 1.121 | 0.2670 | Yes | ||
| 44 | RAB10 | 2870 | 1.112 | 0.2709 | Yes | ||
| 45 | UPF3B | 2881 | 1.110 | 0.2760 | Yes | ||
| 46 | FBXL11 | 3036 | 1.074 | 0.2742 | Yes | ||
| 47 | ZFP37 | 3081 | 1.065 | 0.2775 | Yes | ||
| 48 | PBX3 | 3400 | 0.991 | 0.2679 | No | ||
| 49 | GNAO1 | 3428 | 0.985 | 0.2715 | No | ||
| 50 | RNF26 | 3552 | 0.958 | 0.2706 | No | ||
| 51 | SNAP25 | 3593 | 0.949 | 0.2735 | No | ||
| 52 | AKT1S1 | 3957 | 0.880 | 0.2613 | No | ||
| 53 | WASL | 4168 | 0.843 | 0.2558 | No | ||
| 54 | NTRK3 | 4304 | 0.821 | 0.2537 | No | ||
| 55 | GLG1 | 4365 | 0.809 | 0.2550 | No | ||
| 56 | SFRS3 | 4479 | 0.790 | 0.2537 | No | ||
| 57 | BAD | 4507 | 0.786 | 0.2564 | No | ||
| 58 | MYNN | 4687 | 0.758 | 0.2520 | No | ||
| 59 | MAEA | 4755 | 0.748 | 0.2526 | No | ||
| 60 | TUBA3 | 4974 | 0.712 | 0.2461 | No | ||
| 61 | CNNM1 | 5026 | 0.704 | 0.2473 | No | ||
| 62 | RIF1 | 5417 | 0.646 | 0.2326 | No | ||
| 63 | CSAD | 5608 | 0.621 | 0.2270 | No | ||
| 64 | ARID4A | 5645 | 0.615 | 0.2284 | No | ||
| 65 | EP300 | 5873 | 0.578 | 0.2209 | No | ||
| 66 | CTCF | 5879 | 0.578 | 0.2235 | No | ||
| 67 | RALA | 5982 | 0.562 | 0.2216 | No | ||
| 68 | PUM1 | 6066 | 0.550 | 0.2206 | No | ||
| 69 | WSB1 | 6377 | 0.502 | 0.2088 | No | ||
| 70 | SCRN1 | 6550 | 0.480 | 0.2033 | No | ||
| 71 | RAD21 | 6579 | 0.476 | 0.2044 | No | ||
| 72 | EVI1 | 6748 | 0.452 | 0.1990 | No | ||
| 73 | ERH | 7124 | 0.404 | 0.1838 | No | ||
| 74 | OSBPL11 | 7435 | 0.363 | 0.1713 | No | ||
| 75 | ABCE1 | 7910 | 0.298 | 0.1511 | No | ||
| 76 | UBL3 | 8096 | 0.273 | 0.1439 | No | ||
| 77 | THAP1 | 8360 | 0.238 | 0.1330 | No | ||
| 78 | ZZZ3 | 8870 | 0.166 | 0.1105 | No | ||
| 79 | FASTK | 9101 | 0.132 | 0.1006 | No | ||
| 80 | TAF6 | 9231 | 0.116 | 0.0953 | No | ||
| 81 | DYRK1B | 9653 | 0.051 | 0.0762 | No | ||
| 82 | PSMD8 | 10484 | -0.076 | 0.0385 | No | ||
| 83 | TBC1D17 | 10605 | -0.095 | 0.0334 | No | ||
| 84 | DEAF1 | 10648 | -0.100 | 0.0320 | No | ||
| 85 | MPDU1 | 10676 | -0.104 | 0.0313 | No | ||
| 86 | ATP5F1 | 10909 | -0.143 | 0.0213 | No | ||
| 87 | ADNP | 10963 | -0.150 | 0.0196 | No | ||
| 88 | DHX15 | 10989 | -0.153 | 0.0193 | No | ||
| 89 | PIGL | 11262 | -0.189 | 0.0077 | No | ||
| 90 | RASL11B | 11746 | -0.244 | -0.0132 | No | ||
| 91 | NFYC | 11857 | -0.258 | -0.0170 | No | ||
| 92 | STRN4 | 12114 | -0.285 | -0.0273 | No | ||
| 93 | NACA | 12320 | -0.311 | -0.0352 | No | ||
| 94 | RPL35A | 12550 | -0.339 | -0.0440 | No | ||
| 95 | RPL30 | 12665 | -0.351 | -0.0475 | No | ||
| 96 | BCL2L13 | 13169 | -0.410 | -0.0686 | No | ||
| 97 | PRDM10 | 13184 | -0.411 | -0.0672 | No | ||
| 98 | WDR13 | 13620 | -0.463 | -0.0848 | No | ||
| 99 | USF1 | 13855 | -0.493 | -0.0931 | No | ||
| 100 | BTF3 | 13951 | -0.503 | -0.0950 | No | ||
| 101 | HOXA2 | 13955 | -0.504 | -0.0926 | No | ||
| 102 | PRKCSH | 14468 | -0.565 | -0.1133 | No | ||
| 103 | CDKN2C | 14633 | -0.587 | -0.1179 | No | ||
| 104 | MORF4L2 | 14644 | -0.589 | -0.1154 | No | ||
| 105 | JTB | 14730 | -0.599 | -0.1163 | No | ||
| 106 | FZD8 | 14890 | -0.617 | -0.1206 | No | ||
| 107 | PYY | 15104 | -0.639 | -0.1271 | No | ||
| 108 | ZIC3 | 15674 | -0.712 | -0.1497 | No | ||
| 109 | NUDT3 | 15766 | -0.724 | -0.1503 | No | ||
| 110 | FKRP | 15943 | -0.747 | -0.1547 | No | ||
| 111 | RAB1B | 15974 | -0.753 | -0.1523 | No | ||
| 112 | RASGRP2 | 16042 | -0.763 | -0.1516 | No | ||
| 113 | MARK3 | 16118 | -0.773 | -0.1511 | No | ||
| 114 | ARF1 | 16466 | -0.818 | -0.1630 | No | ||
| 115 | B3GALT6 | 16641 | -0.840 | -0.1668 | No | ||
| 116 | DGCR2 | 16810 | -0.863 | -0.1702 | No | ||
| 117 | E2F6 | 17484 | -0.969 | -0.1963 | No | ||
| 118 | NASP | 17539 | -0.979 | -0.1939 | No | ||
| 119 | MGAT4B | 17784 | -1.019 | -0.2000 | No | ||
| 120 | AP4S1 | 17877 | -1.038 | -0.1991 | No | ||
| 121 | SART3 | 18091 | -1.074 | -0.2035 | No | ||
| 122 | HIC2 | 18205 | -1.093 | -0.2032 | No | ||
| 123 | SMYD5 | 18225 | -1.098 | -0.1986 | No | ||
| 124 | LYPLA2 | 18325 | -1.120 | -0.1976 | No | ||
| 125 | ADK | 18350 | -1.124 | -0.1931 | No | ||
| 126 | KERA | 18585 | -1.171 | -0.1980 | No | ||
| 127 | TBC1D14 | 18653 | -1.185 | -0.1952 | No | ||
| 128 | ACTR8 | 18665 | -1.187 | -0.1898 | No | ||
| 129 | EIF4G1 | 18692 | -1.193 | -0.1850 | No | ||
| 130 | CNOT3 | 18921 | -1.245 | -0.1893 | No | ||
| 131 | ABHD1 | 18960 | -1.253 | -0.1848 | No | ||
| 132 | AIP | 19257 | -1.325 | -0.1918 | No | ||
| 133 | PIAS3 | 19406 | -1.366 | -0.1918 | No | ||
| 134 | TOP3A | 19429 | -1.373 | -0.1860 | No | ||
| 135 | GTF3C2 | 19451 | -1.378 | -0.1801 | No | ||
| 136 | TSC1 | 19466 | -1.385 | -0.1738 | No | ||
| 137 | AP3D1 | 19485 | -1.390 | -0.1677 | No | ||
| 138 | SYNGR1 | 19615 | -1.430 | -0.1665 | No | ||
| 139 | ZIM2 | 19623 | -1.432 | -0.1597 | No | ||
| 140 | SMCR8 | 19625 | -1.433 | -0.1526 | No | ||
| 141 | DAP3 | 19633 | -1.434 | -0.1458 | No | ||
| 142 | CCND1 | 19763 | -1.482 | -0.1443 | No | ||
| 143 | NCDN | 19901 | -1.528 | -0.1430 | No | ||
| 144 | CCNK | 19974 | -1.555 | -0.1386 | No | ||
| 145 | AP1G1 | 20050 | -1.590 | -0.1341 | No | ||
| 146 | SELM | 20166 | -1.637 | -0.1312 | No | ||
| 147 | ODF2 | 20421 | -1.750 | -0.1342 | No | ||
| 148 | ENSA | 20479 | -1.781 | -0.1279 | No | ||
| 149 | H1F0 | 20504 | -1.798 | -0.1201 | No | ||
| 150 | FBXO9 | 20550 | -1.821 | -0.1131 | No | ||
| 151 | FUT10 | 20748 | -1.934 | -0.1125 | No | ||
| 152 | RUFY1 | 21008 | -2.121 | -0.1138 | No | ||
| 153 | POLR3E | 21059 | -2.154 | -0.1054 | No | ||
| 154 | ASB2 | 21165 | -2.252 | -0.0990 | No | ||
| 155 | CAMLG | 21177 | -2.270 | -0.0882 | No | ||
| 156 | DZIP1 | 21203 | -2.295 | -0.0779 | No | ||
| 157 | NDRG3 | 21206 | -2.296 | -0.0665 | No | ||
| 158 | UBTF | 21212 | -2.303 | -0.0553 | No | ||
| 159 | MAP3K4 | 21340 | -2.470 | -0.0488 | No | ||
| 160 | GBF1 | 21377 | -2.528 | -0.0379 | No | ||
| 161 | VASP | 21648 | -3.238 | -0.0342 | No | ||
| 162 | IER2 | 21848 | -4.749 | -0.0197 | No | ||
| 163 | PCBP4 | 21856 | -4.849 | 0.0042 | No |