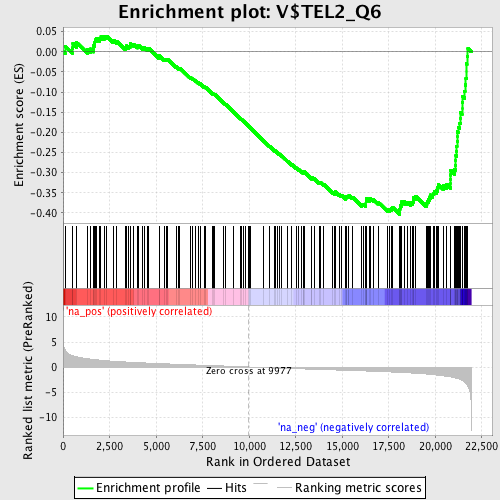
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$TEL2_Q6 |
| Enrichment Score (ES) | -0.4040005 |
| Normalized Enrichment Score (NES) | -1.7710677 |
| Nominal p-value | 0.0013679891 |
| FDR q-value | 0.016731763 |
| FWER p-Value | 0.189 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TCF12 | 109 | 3.364 | 0.0140 | No | ||
| 2 | DAPP1 | 486 | 2.292 | 0.0096 | No | ||
| 3 | HERC4 | 527 | 2.241 | 0.0205 | No | ||
| 4 | USP3 | 729 | 2.028 | 0.0227 | No | ||
| 5 | HMGA1 | 1301 | 1.652 | 0.0058 | No | ||
| 6 | CHES1 | 1467 | 1.573 | 0.0071 | No | ||
| 7 | DMTF1 | 1633 | 1.505 | 0.0080 | No | ||
| 8 | PRKACB | 1648 | 1.500 | 0.0158 | No | ||
| 9 | RNF12 | 1689 | 1.488 | 0.0224 | No | ||
| 10 | ERCC6 | 1713 | 1.476 | 0.0297 | No | ||
| 11 | CANX | 1790 | 1.442 | 0.0343 | No | ||
| 12 | CHD2 | 1962 | 1.372 | 0.0342 | No | ||
| 13 | ARHGAP15 | 2034 | 1.350 | 0.0386 | No | ||
| 14 | DPPA4 | 2201 | 1.295 | 0.0382 | No | ||
| 15 | NR4A2 | 2339 | 1.246 | 0.0390 | No | ||
| 16 | PPP4C | 2692 | 1.153 | 0.0293 | No | ||
| 17 | HOXC4 | 2893 | 1.107 | 0.0264 | No | ||
| 18 | CAST | 3358 | 0.999 | 0.0107 | No | ||
| 19 | CLMN | 3393 | 0.992 | 0.0148 | No | ||
| 20 | MGAT4A | 3524 | 0.964 | 0.0143 | No | ||
| 21 | HM13 | 3596 | 0.949 | 0.0163 | No | ||
| 22 | RNF6 | 3600 | 0.948 | 0.0216 | No | ||
| 23 | SPIB | 3763 | 0.916 | 0.0193 | No | ||
| 24 | CAPZA1 | 3978 | 0.876 | 0.0144 | No | ||
| 25 | CKS1B | 4061 | 0.861 | 0.0155 | No | ||
| 26 | DDX50 | 4283 | 0.824 | 0.0100 | No | ||
| 27 | DCLRE1C | 4362 | 0.809 | 0.0110 | No | ||
| 28 | BTAF1 | 4531 | 0.781 | 0.0077 | No | ||
| 29 | CGGBP1 | 4612 | 0.770 | 0.0084 | No | ||
| 30 | NXT2 | 5165 | 0.686 | -0.0131 | No | ||
| 31 | UGCGL2 | 5167 | 0.685 | -0.0093 | No | ||
| 32 | TRIM41 | 5435 | 0.644 | -0.0179 | No | ||
| 33 | FXC1 | 5541 | 0.629 | -0.0192 | No | ||
| 34 | CSAD | 5608 | 0.621 | -0.0187 | No | ||
| 35 | TMEM24 | 6068 | 0.550 | -0.0366 | No | ||
| 36 | SIRT3 | 6211 | 0.525 | -0.0402 | No | ||
| 37 | AGPAT1 | 6276 | 0.515 | -0.0402 | No | ||
| 38 | C1QTNF6 | 6835 | 0.440 | -0.0634 | No | ||
| 39 | AGL | 6973 | 0.423 | -0.0673 | No | ||
| 40 | ERH | 7124 | 0.404 | -0.0719 | No | ||
| 41 | SNRPB | 7269 | 0.385 | -0.0763 | No | ||
| 42 | FIBP | 7382 | 0.369 | -0.0794 | No | ||
| 43 | STAT4 | 7591 | 0.341 | -0.0870 | No | ||
| 44 | SOX14 | 7656 | 0.331 | -0.0880 | No | ||
| 45 | SLC9A3R1 | 8031 | 0.284 | -0.1036 | No | ||
| 46 | AMD1 | 8106 | 0.272 | -0.1055 | No | ||
| 47 | U2AF2 | 8138 | 0.268 | -0.1054 | No | ||
| 48 | GGTL3 | 8616 | 0.202 | -0.1261 | No | ||
| 49 | IL23A | 8716 | 0.188 | -0.1296 | No | ||
| 50 | TAF5 | 9168 | 0.124 | -0.1496 | No | ||
| 51 | DIABLO | 9521 | 0.071 | -0.1654 | No | ||
| 52 | PPIL1 | 9589 | 0.062 | -0.1681 | No | ||
| 53 | POU3F4 | 9717 | 0.041 | -0.1737 | No | ||
| 54 | RHOV | 9784 | 0.030 | -0.1765 | No | ||
| 55 | SLC39A11 | 9958 | 0.003 | -0.1845 | No | ||
| 56 | SEMA4C | 10003 | -0.005 | -0.1864 | No | ||
| 57 | STARD13 | 10083 | -0.016 | -0.1900 | No | ||
| 58 | OGG1 | 10786 | -0.122 | -0.2215 | No | ||
| 59 | VAMP8 | 11082 | -0.166 | -0.2341 | No | ||
| 60 | TEAD3 | 11332 | -0.198 | -0.2444 | No | ||
| 61 | TRAPPC1 | 11391 | -0.204 | -0.2459 | No | ||
| 62 | TBN | 11503 | -0.217 | -0.2498 | No | ||
| 63 | GFI1 | 11618 | -0.230 | -0.2537 | No | ||
| 64 | NR1D1 | 11759 | -0.246 | -0.2588 | No | ||
| 65 | HYAL2 | 12082 | -0.281 | -0.2720 | No | ||
| 66 | PSMD13 | 12297 | -0.309 | -0.2800 | No | ||
| 67 | DDIT3 | 12521 | -0.335 | -0.2884 | No | ||
| 68 | LIN28 | 12659 | -0.351 | -0.2927 | No | ||
| 69 | TIMM10 | 12824 | -0.370 | -0.2981 | No | ||
| 70 | FCHO1 | 12898 | -0.379 | -0.2993 | No | ||
| 71 | TENC1 | 12903 | -0.380 | -0.2974 | No | ||
| 72 | CRSP7 | 12956 | -0.385 | -0.2976 | No | ||
| 73 | ING4 | 13361 | -0.434 | -0.3137 | No | ||
| 74 | ADAMTS4 | 13367 | -0.435 | -0.3115 | No | ||
| 75 | MAP3K11 | 13487 | -0.449 | -0.3144 | No | ||
| 76 | GRB7 | 13779 | -0.484 | -0.3250 | No | ||
| 77 | ELK4 | 13831 | -0.491 | -0.3246 | No | ||
| 78 | JPH4 | 13985 | -0.508 | -0.3287 | No | ||
| 79 | ARFIP2 | 14492 | -0.569 | -0.3488 | No | ||
| 80 | FBXO22 | 14604 | -0.582 | -0.3506 | No | ||
| 81 | POLL | 14613 | -0.584 | -0.3476 | No | ||
| 82 | CENTD2 | 14838 | -0.611 | -0.3545 | No | ||
| 83 | DES | 14963 | -0.625 | -0.3566 | No | ||
| 84 | MUSK | 15198 | -0.652 | -0.3637 | No | ||
| 85 | RGS14 | 15203 | -0.653 | -0.3602 | No | ||
| 86 | ACSL5 | 15243 | -0.656 | -0.3583 | No | ||
| 87 | UBE2N | 15327 | -0.668 | -0.3583 | No | ||
| 88 | NDUFS2 | 15358 | -0.672 | -0.3559 | No | ||
| 89 | PSMC2 | 15560 | -0.695 | -0.3612 | No | ||
| 90 | CD2BP2 | 16051 | -0.764 | -0.3794 | No | ||
| 91 | MARK3 | 16118 | -0.773 | -0.3781 | No | ||
| 92 | RIN1 | 16270 | -0.793 | -0.3805 | No | ||
| 93 | RNPS1 | 16272 | -0.793 | -0.3761 | No | ||
| 94 | NEDD8 | 16274 | -0.793 | -0.3717 | No | ||
| 95 | CASP8 | 16284 | -0.795 | -0.3676 | No | ||
| 96 | PNMA1 | 16302 | -0.798 | -0.3639 | No | ||
| 97 | CD3E | 16458 | -0.818 | -0.3664 | No | ||
| 98 | LTBR | 16514 | -0.825 | -0.3642 | No | ||
| 99 | EXTL2 | 16672 | -0.845 | -0.3667 | No | ||
| 100 | ZFP95 | 16922 | -0.880 | -0.3731 | No | ||
| 101 | ST7L | 17434 | -0.959 | -0.3912 | No | ||
| 102 | NASP | 17539 | -0.979 | -0.3904 | No | ||
| 103 | PRKACA | 17631 | -0.996 | -0.3890 | No | ||
| 104 | HIRIP3 | 17702 | -1.008 | -0.3865 | No | ||
| 105 | DGKA | 18084 | -1.073 | -0.3979 | Yes | ||
| 106 | SART3 | 18091 | -1.074 | -0.3922 | Yes | ||
| 107 | SHC1 | 18103 | -1.076 | -0.3866 | Yes | ||
| 108 | MEIS1 | 18116 | -1.078 | -0.3811 | Yes | ||
| 109 | PAFAH1B2 | 18172 | -1.087 | -0.3775 | Yes | ||
| 110 | PLCG1 | 18203 | -1.093 | -0.3727 | Yes | ||
| 111 | LYPLA2 | 18325 | -1.120 | -0.3719 | Yes | ||
| 112 | E2F5 | 18509 | -1.155 | -0.3738 | Yes | ||
| 113 | CPNE8 | 18687 | -1.192 | -0.3752 | Yes | ||
| 114 | HTATIP | 18792 | -1.215 | -0.3731 | Yes | ||
| 115 | CCL2 | 18809 | -1.219 | -0.3670 | Yes | ||
| 116 | ZDHHC5 | 18829 | -1.223 | -0.3609 | Yes | ||
| 117 | CENTG1 | 18945 | -1.250 | -0.3592 | Yes | ||
| 118 | LCP1 | 19513 | -1.397 | -0.3773 | Yes | ||
| 119 | DPP3 | 19553 | -1.411 | -0.3711 | Yes | ||
| 120 | SNX1 | 19641 | -1.437 | -0.3670 | Yes | ||
| 121 | SCAMP2 | 19680 | -1.452 | -0.3606 | Yes | ||
| 122 | PTK2 | 19751 | -1.476 | -0.3555 | Yes | ||
| 123 | FURIN | 19875 | -1.519 | -0.3525 | Yes | ||
| 124 | HCLS1 | 19929 | -1.540 | -0.3463 | Yes | ||
| 125 | TUSC3 | 20075 | -1.600 | -0.3439 | Yes | ||
| 126 | IKBKB | 20132 | -1.623 | -0.3373 | Yes | ||
| 127 | BIN3 | 20153 | -1.630 | -0.3290 | Yes | ||
| 128 | MRPL52 | 20424 | -1.751 | -0.3316 | Yes | ||
| 129 | ESRRA | 20618 | -1.856 | -0.3299 | Yes | ||
| 130 | FXYD5 | 20809 | -1.973 | -0.3275 | Yes | ||
| 131 | ARHGAP4 | 20814 | -1.975 | -0.3166 | Yes | ||
| 132 | RALB | 20823 | -1.981 | -0.3058 | Yes | ||
| 133 | CAP1 | 20831 | -1.986 | -0.2949 | Yes | ||
| 134 | PRKCBP1 | 21031 | -2.135 | -0.2920 | Yes | ||
| 135 | STX11 | 21074 | -2.169 | -0.2817 | Yes | ||
| 136 | ARHGAP1 | 21097 | -2.192 | -0.2703 | Yes | ||
| 137 | MARK1 | 21105 | -2.199 | -0.2582 | Yes | ||
| 138 | DNAJC14 | 21135 | -2.225 | -0.2470 | Yes | ||
| 139 | MAP4K2 | 21147 | -2.232 | -0.2349 | Yes | ||
| 140 | TGIF2 | 21166 | -2.254 | -0.2230 | Yes | ||
| 141 | ARHGEF7 | 21179 | -2.271 | -0.2108 | Yes | ||
| 142 | CTSS | 21186 | -2.279 | -0.1982 | Yes | ||
| 143 | ARRB2 | 21227 | -2.318 | -0.1870 | Yes | ||
| 144 | PIK3R4 | 21313 | -2.435 | -0.1771 | Yes | ||
| 145 | SIPA1 | 21344 | -2.478 | -0.1645 | Yes | ||
| 146 | SLC25A5 | 21353 | -2.489 | -0.1508 | Yes | ||
| 147 | RIN3 | 21437 | -2.664 | -0.1396 | Yes | ||
| 148 | JUNB | 21440 | -2.670 | -0.1247 | Yes | ||
| 149 | SEC24C | 21465 | -2.711 | -0.1105 | Yes | ||
| 150 | SLC30A7 | 21579 | -3.016 | -0.0986 | Yes | ||
| 151 | PTPN6 | 21597 | -3.085 | -0.0820 | Yes | ||
| 152 | GMPR2 | 21644 | -3.221 | -0.0660 | Yes | ||
| 153 | CD79A | 21671 | -3.365 | -0.0482 | Yes | ||
| 154 | CTSW | 21672 | -3.373 | -0.0291 | Yes | ||
| 155 | CORO1C | 21703 | -3.548 | -0.0105 | Yes | ||
| 156 | RGS3 | 21752 | -3.840 | 0.0089 | Yes |