Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$TCF11MAFG_01 |
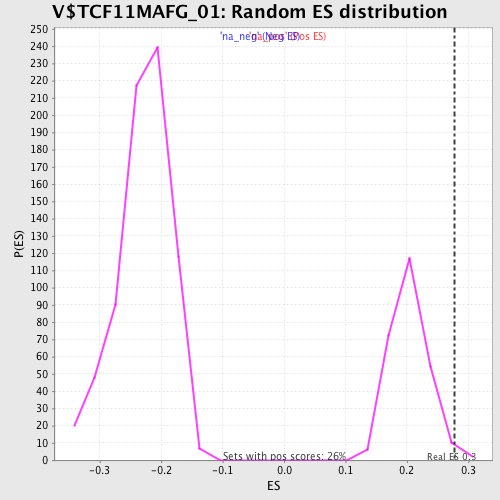
| Enrichment Score (ES) | 0.27802885 |
| Normalized Enrichment Score (NES) | 1.3642944 |
| Nominal p-value | 0.019157087 |
| FDR q-value | 0.21900758 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BIRC6 | 177 | 2.985 | 0.0114 | Yes | ||
| 2 | USP14 | 182 | 2.980 | 0.0308 | Yes | ||
| 3 | IGSF4 | 212 | 2.865 | 0.0482 | Yes | ||
| 4 | RTN3 | 317 | 2.572 | 0.0603 | Yes | ||
| 5 | RABEP1 | 443 | 2.362 | 0.0700 | Yes | ||
| 6 | TBL1X | 480 | 2.303 | 0.0835 | Yes | ||
| 7 | PAFAH1B1 | 513 | 2.259 | 0.0968 | Yes | ||
| 8 | PPP2CA | 652 | 2.103 | 0.1042 | Yes | ||
| 9 | JARID2 | 776 | 1.986 | 0.1116 | Yes | ||
| 10 | TLK1 | 843 | 1.923 | 0.1212 | Yes | ||
| 11 | ANGPT1 | 1254 | 1.681 | 0.1134 | Yes | ||
| 12 | PCYT1A | 1378 | 1.614 | 0.1183 | Yes | ||
| 13 | KCNA2 | 1423 | 1.595 | 0.1267 | Yes | ||
| 14 | TIAL1 | 1439 | 1.589 | 0.1365 | Yes | ||
| 15 | FOXP1 | 1454 | 1.579 | 0.1462 | Yes | ||
| 16 | SPRED1 | 1466 | 1.574 | 0.1560 | Yes | ||
| 17 | GAB2 | 1486 | 1.568 | 0.1654 | Yes | ||
| 18 | ATP1B1 | 1557 | 1.539 | 0.1722 | Yes | ||
| 19 | PSMA6 | 1651 | 1.499 | 0.1778 | Yes | ||
| 20 | FLI1 | 1719 | 1.474 | 0.1844 | Yes | ||
| 21 | PPARGC1A | 1723 | 1.472 | 0.1939 | Yes | ||
| 22 | RAB1A | 1738 | 1.468 | 0.2028 | Yes | ||
| 23 | WDFY3 | 2082 | 1.334 | 0.1959 | Yes | ||
| 24 | MITF | 2097 | 1.330 | 0.2039 | Yes | ||
| 25 | TAC1 | 2214 | 1.291 | 0.2071 | Yes | ||
| 26 | NPEPPS | 2221 | 1.287 | 0.2152 | Yes | ||
| 27 | MDM2 | 2278 | 1.267 | 0.2209 | Yes | ||
| 28 | TOMM70A | 2307 | 1.257 | 0.2279 | Yes | ||
| 29 | NUMBL | 2356 | 1.240 | 0.2338 | Yes | ||
| 30 | PTP4A1 | 2359 | 1.239 | 0.2418 | Yes | ||
| 31 | SOX5 | 2456 | 1.209 | 0.2454 | Yes | ||
| 32 | GAP43 | 2480 | 1.203 | 0.2522 | Yes | ||
| 33 | GTF2A1 | 2485 | 1.202 | 0.2599 | Yes | ||
| 34 | MLL3 | 2693 | 1.153 | 0.2579 | Yes | ||
| 35 | NUDT11 | 2799 | 1.127 | 0.2605 | Yes | ||
| 36 | RAB10 | 2870 | 1.112 | 0.2646 | Yes | ||
| 37 | LRP1B | 2971 | 1.090 | 0.2671 | Yes | ||
| 38 | CDC45L | 3011 | 1.081 | 0.2724 | Yes | ||
| 39 | ATP6V0A1 | 3165 | 1.045 | 0.2722 | Yes | ||
| 40 | FBXO30 | 3254 | 1.024 | 0.2749 | Yes | ||
| 41 | SERPINB5 | 3375 | 0.996 | 0.2759 | Yes | ||
| 42 | CRYGS | 3545 | 0.960 | 0.2745 | Yes | ||
| 43 | NRXN2 | 3797 | 0.909 | 0.2689 | Yes | ||
| 44 | SFXN5 | 3897 | 0.892 | 0.2702 | Yes | ||
| 45 | AKT1S1 | 3957 | 0.880 | 0.2733 | Yes | ||
| 46 | SATB1 | 3979 | 0.876 | 0.2780 | Yes | ||
| 47 | ERO1L | 4307 | 0.820 | 0.2684 | No | ||
| 48 | ARHGEF11 | 4378 | 0.807 | 0.2705 | No | ||
| 49 | GPX2 | 4490 | 0.788 | 0.2705 | No | ||
| 50 | ARPP-19 | 4801 | 0.739 | 0.2612 | No | ||
| 51 | IRX4 | 4852 | 0.732 | 0.2637 | No | ||
| 52 | IL13RA1 | 4859 | 0.730 | 0.2682 | No | ||
| 53 | RIMS2 | 5217 | 0.677 | 0.2562 | No | ||
| 54 | LRRFIP2 | 5218 | 0.677 | 0.2607 | No | ||
| 55 | ST5 | 5257 | 0.672 | 0.2633 | No | ||
| 56 | EIF5 | 5447 | 0.642 | 0.2589 | No | ||
| 57 | FGF12 | 5464 | 0.640 | 0.2623 | No | ||
| 58 | NUDT10 | 5482 | 0.637 | 0.2657 | No | ||
| 59 | FLNC | 5586 | 0.624 | 0.2651 | No | ||
| 60 | SLC11A1 | 5596 | 0.623 | 0.2687 | No | ||
| 61 | SV2B | 6035 | 0.553 | 0.2523 | No | ||
| 62 | SNCB | 6345 | 0.507 | 0.2414 | No | ||
| 63 | KIRREL3 | 6730 | 0.455 | 0.2268 | No | ||
| 64 | DRP2 | 6762 | 0.450 | 0.2283 | No | ||
| 65 | TLL1 | 6862 | 0.436 | 0.2266 | No | ||
| 66 | E2F3 | 7113 | 0.406 | 0.2178 | No | ||
| 67 | LRRTM4 | 7211 | 0.393 | 0.2159 | No | ||
| 68 | CST7 | 7620 | 0.337 | 0.1994 | No | ||
| 69 | KCNE4 | 7744 | 0.321 | 0.1959 | No | ||
| 70 | ASPSCR1 | 7927 | 0.297 | 0.1895 | No | ||
| 71 | IFNB1 | 8083 | 0.276 | 0.1842 | No | ||
| 72 | SLC6A5 | 8217 | 0.259 | 0.1798 | No | ||
| 73 | PPARG | 8381 | 0.235 | 0.1738 | No | ||
| 74 | APBA2BP | 8633 | 0.200 | 0.1636 | No | ||
| 75 | P2RXL1 | 9017 | 0.144 | 0.1470 | No | ||
| 76 | PGRMC1 | 9681 | 0.046 | 0.1169 | No | ||
| 77 | BTK | 9783 | 0.031 | 0.1125 | No | ||
| 78 | SKP1A | 10018 | -0.007 | 0.1018 | No | ||
| 79 | POPDC3 | 10405 | -0.066 | 0.0845 | No | ||
| 80 | WNT3 | 10416 | -0.067 | 0.0845 | No | ||
| 81 | SPTBN4 | 10582 | -0.091 | 0.0775 | No | ||
| 82 | TBC1D17 | 10605 | -0.095 | 0.0771 | No | ||
| 83 | BLVRB | 10612 | -0.096 | 0.0775 | No | ||
| 84 | PCDHA10 | 10977 | -0.152 | 0.0618 | No | ||
| 85 | PLS3 | 11273 | -0.190 | 0.0495 | No | ||
| 86 | BEX2 | 11375 | -0.202 | 0.0462 | No | ||
| 87 | NEUROD6 | 11432 | -0.208 | 0.0450 | No | ||
| 88 | IPO13 | 11583 | -0.226 | 0.0396 | No | ||
| 89 | SLC6A4 | 11680 | -0.237 | 0.0367 | No | ||
| 90 | NLGN2 | 12078 | -0.281 | 0.0203 | No | ||
| 91 | MRC2 | 12259 | -0.302 | 0.0140 | No | ||
| 92 | MRPL32 | 12284 | -0.307 | 0.0150 | No | ||
| 93 | TUBB | 12290 | -0.308 | 0.0167 | No | ||
| 94 | ABCB6 | 12927 | -0.383 | -0.0099 | No | ||
| 95 | TINAG | 13010 | -0.392 | -0.0111 | No | ||
| 96 | SPATS2 | 13283 | -0.423 | -0.0208 | No | ||
| 97 | FGF9 | 13392 | -0.438 | -0.0229 | No | ||
| 98 | HSPB3 | 13442 | -0.445 | -0.0222 | No | ||
| 99 | PRDX1 | 13722 | -0.476 | -0.0319 | No | ||
| 100 | PPP2R2C | 13969 | -0.505 | -0.0399 | No | ||
| 101 | DUSP13 | 14250 | -0.539 | -0.0492 | No | ||
| 102 | LAMC1 | 14510 | -0.571 | -0.0574 | No | ||
| 103 | HSPB8 | 14623 | -0.586 | -0.0587 | No | ||
| 104 | TFEC | 15308 | -0.666 | -0.0857 | No | ||
| 105 | OMG | 15984 | -0.754 | -0.1117 | No | ||
| 106 | IL6 | 16016 | -0.760 | -0.1081 | No | ||
| 107 | TXNRD1 | 16140 | -0.775 | -0.1087 | No | ||
| 108 | FOSL1 | 16325 | -0.800 | -0.1119 | No | ||
| 109 | PCDH9 | 16469 | -0.819 | -0.1131 | No | ||
| 110 | UCHL1 | 16522 | -0.826 | -0.1101 | No | ||
| 111 | RBBP7 | 16538 | -0.828 | -0.1053 | No | ||
| 112 | BRD2 | 16659 | -0.843 | -0.1053 | No | ||
| 113 | LOXL4 | 16846 | -0.868 | -0.1082 | No | ||
| 114 | CBX6 | 16868 | -0.871 | -0.1034 | No | ||
| 115 | SYT2 | 16878 | -0.872 | -0.0981 | No | ||
| 116 | PSMD11 | 16972 | -0.887 | -0.0966 | No | ||
| 117 | PSMA5 | 16974 | -0.887 | -0.0908 | No | ||
| 118 | RASGRF1 | 17064 | -0.903 | -0.0890 | No | ||
| 119 | GRIK2 | 17154 | -0.916 | -0.0871 | No | ||
| 120 | PFN2 | 17223 | -0.926 | -0.0841 | No | ||
| 121 | PDHA2 | 17378 | -0.949 | -0.0850 | No | ||
| 122 | OSBPL5 | 17530 | -0.978 | -0.0855 | No | ||
| 123 | ITPK1 | 17548 | -0.981 | -0.0799 | No | ||
| 124 | HECTD2 | 18082 | -1.073 | -0.0973 | No | ||
| 125 | PDGFB | 18137 | -1.080 | -0.0927 | No | ||
| 126 | RORB | 18148 | -1.084 | -0.0860 | No | ||
| 127 | UFD1L | 18198 | -1.091 | -0.0811 | No | ||
| 128 | PSMA2 | 18226 | -1.098 | -0.0752 | No | ||
| 129 | A2BP1 | 18581 | -1.170 | -0.0838 | No | ||
| 130 | FIBCD1 | 18584 | -1.171 | -0.0762 | No | ||
| 131 | MGST1 | 18670 | -1.188 | -0.0723 | No | ||
| 132 | EIF4G1 | 18692 | -1.193 | -0.0654 | No | ||
| 133 | LEPROTL1 | 18813 | -1.221 | -0.0630 | No | ||
| 134 | GRK6 | 19126 | -1.289 | -0.0688 | No | ||
| 135 | CALM3 | 19591 | -1.421 | -0.0808 | No | ||
| 136 | SNX12 | 19734 | -1.473 | -0.0777 | No | ||
| 137 | ALDOA | 19736 | -1.473 | -0.0681 | No | ||
| 138 | ZIC4 | 20073 | -1.599 | -0.0730 | No | ||
| 139 | ATP2A3 | 20235 | -1.669 | -0.0695 | No | ||
| 140 | UBE4B | 20303 | -1.700 | -0.0614 | No | ||
| 141 | MNT | 20472 | -1.778 | -0.0575 | No | ||
| 142 | DMD | 20507 | -1.799 | -0.0472 | No | ||
| 143 | LIMK1 | 20601 | -1.848 | -0.0394 | No | ||
| 144 | CD69 | 20802 | -1.970 | -0.0357 | No | ||
| 145 | PBX2 | 20850 | -1.998 | -0.0247 | No | ||
| 146 | HSD11B1 | 21043 | -2.148 | -0.0195 | No | ||
| 147 | ITM2B | 21113 | -2.204 | -0.0082 | No | ||
| 148 | LMO4 | 21122 | -2.212 | 0.0059 | No | ||
| 149 | ASB2 | 21165 | -2.252 | 0.0188 | No | ||
| 150 | ENO1 | 21416 | -2.605 | 0.0244 | No |