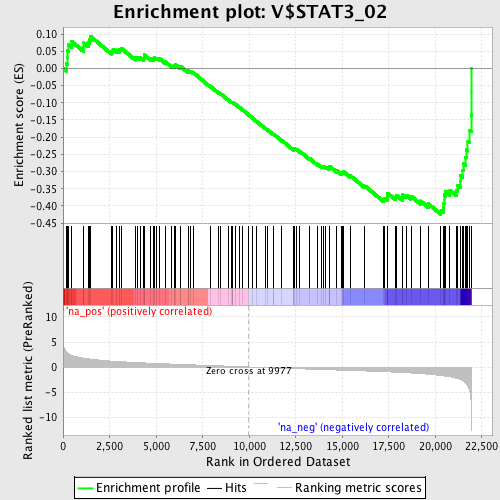
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$STAT3_02 |
| Enrichment Score (ES) | -0.42488998 |
| Normalized Enrichment Score (NES) | -1.7408206 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.017368838 |
| FWER p-Value | 0.259 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TCF7L2 | 168 | 3.009 | 0.0144 | No | ||
| 2 | FUT8 | 246 | 2.749 | 0.0311 | No | ||
| 3 | SOCS1 | 249 | 2.736 | 0.0510 | No | ||
| 4 | AP2B1 | 273 | 2.682 | 0.0697 | No | ||
| 5 | COL4A3BP | 445 | 2.359 | 0.0792 | No | ||
| 6 | NAPB | 1116 | 1.749 | 0.0613 | No | ||
| 7 | EIF4E | 1119 | 1.747 | 0.0741 | No | ||
| 8 | NCAM1 | 1348 | 1.630 | 0.0756 | No | ||
| 9 | GPHN | 1432 | 1.593 | 0.0835 | No | ||
| 10 | LRP2 | 1457 | 1.579 | 0.0940 | No | ||
| 11 | SLC35A5 | 2598 | 1.177 | 0.0504 | No | ||
| 12 | SDC1 | 2679 | 1.157 | 0.0553 | No | ||
| 13 | HOXC4 | 2893 | 1.107 | 0.0536 | No | ||
| 14 | CTGF | 3039 | 1.073 | 0.0549 | No | ||
| 15 | CENTD1 | 3126 | 1.054 | 0.0587 | No | ||
| 16 | ARX | 3874 | 0.895 | 0.0310 | No | ||
| 17 | CAPZA1 | 3978 | 0.876 | 0.0327 | No | ||
| 18 | AP1S2 | 4155 | 0.845 | 0.0309 | No | ||
| 19 | KPNB1 | 4321 | 0.818 | 0.0293 | No | ||
| 20 | LTBP1 | 4349 | 0.813 | 0.0341 | No | ||
| 21 | GPC3 | 4358 | 0.810 | 0.0397 | No | ||
| 22 | KCNH3 | 4721 | 0.753 | 0.0286 | No | ||
| 23 | CPA4 | 4862 | 0.730 | 0.0276 | No | ||
| 24 | RBPSUH | 4891 | 0.726 | 0.0316 | No | ||
| 25 | RIMS1 | 5039 | 0.702 | 0.0300 | No | ||
| 26 | MIS12 | 5171 | 0.685 | 0.0291 | No | ||
| 27 | EIF5A | 5477 | 0.638 | 0.0198 | No | ||
| 28 | HOXB4 | 5849 | 0.581 | 0.0071 | No | ||
| 29 | BMI1 | 5972 | 0.563 | 0.0056 | No | ||
| 30 | ZBTB11 | 5985 | 0.561 | 0.0092 | No | ||
| 31 | SV2B | 6035 | 0.553 | 0.0110 | No | ||
| 32 | ZFYVE9 | 6282 | 0.514 | 0.0035 | No | ||
| 33 | MOBKL2C | 6294 | 0.513 | 0.0068 | No | ||
| 34 | PPFIA2 | 6718 | 0.457 | -0.0092 | No | ||
| 35 | KIRREL3 | 6730 | 0.455 | -0.0064 | No | ||
| 36 | SPON1 | 6847 | 0.438 | -0.0085 | No | ||
| 37 | SET | 6988 | 0.421 | -0.0118 | No | ||
| 38 | FOSB | 7935 | 0.296 | -0.0530 | No | ||
| 39 | FLRT1 | 8357 | 0.238 | -0.0705 | No | ||
| 40 | KCNN2 | 8469 | 0.222 | -0.0739 | No | ||
| 41 | BMP4 | 8880 | 0.165 | -0.0915 | No | ||
| 42 | UPK2 | 9064 | 0.137 | -0.0989 | No | ||
| 43 | ACCN1 | 9077 | 0.135 | -0.0984 | No | ||
| 44 | FBN2 | 9096 | 0.133 | -0.0983 | No | ||
| 45 | HOXB9 | 9119 | 0.131 | -0.0983 | No | ||
| 46 | RBPSUHL | 9256 | 0.112 | -0.1037 | No | ||
| 47 | ARHGAP8 | 9477 | 0.078 | -0.1132 | No | ||
| 48 | BNC1 | 9647 | 0.051 | -0.1206 | No | ||
| 49 | PAPD1 | 9946 | 0.004 | -0.1342 | No | ||
| 50 | ADM2 | 10164 | -0.028 | -0.1439 | No | ||
| 51 | MLLT7 | 10380 | -0.062 | -0.1533 | No | ||
| 52 | S100A14 | 10851 | -0.132 | -0.1739 | No | ||
| 53 | NELL2 | 10988 | -0.153 | -0.1790 | No | ||
| 54 | CALU | 11279 | -0.191 | -0.1909 | No | ||
| 55 | MATN4 | 11320 | -0.196 | -0.1913 | No | ||
| 56 | NR1D1 | 11759 | -0.246 | -0.2095 | No | ||
| 57 | SLCO5A1 | 12383 | -0.318 | -0.2357 | No | ||
| 58 | PGF | 12393 | -0.319 | -0.2338 | No | ||
| 59 | TRIP10 | 12447 | -0.325 | -0.2338 | No | ||
| 60 | DDIT3 | 12521 | -0.335 | -0.2347 | No | ||
| 61 | WEE1 | 12721 | -0.358 | -0.2412 | No | ||
| 62 | KAZALD1 | 13241 | -0.417 | -0.2619 | No | ||
| 63 | CHRM1 | 13669 | -0.470 | -0.2780 | No | ||
| 64 | WNT4 | 13890 | -0.497 | -0.2844 | No | ||
| 65 | SENP3 | 14000 | -0.509 | -0.2857 | No | ||
| 66 | HOXC6 | 14117 | -0.521 | -0.2871 | No | ||
| 67 | IL18BP | 14288 | -0.543 | -0.2909 | No | ||
| 68 | YY1 | 14290 | -0.543 | -0.2870 | No | ||
| 69 | RND1 | 14328 | -0.548 | -0.2847 | No | ||
| 70 | MYT1 | 14709 | -0.597 | -0.2977 | No | ||
| 71 | GRIN2D | 14936 | -0.622 | -0.3035 | No | ||
| 72 | IRX5 | 15000 | -0.628 | -0.3017 | No | ||
| 73 | HEYL | 15079 | -0.637 | -0.3006 | No | ||
| 74 | CRTAC1 | 15450 | -0.683 | -0.3126 | No | ||
| 75 | CPLX2 | 16219 | -0.786 | -0.3419 | No | ||
| 76 | BCL7A | 17212 | -0.925 | -0.3806 | No | ||
| 77 | NDST2 | 17285 | -0.935 | -0.3770 | No | ||
| 78 | HOXB13 | 17418 | -0.957 | -0.3760 | No | ||
| 79 | MBD6 | 17426 | -0.958 | -0.3693 | No | ||
| 80 | ST7L | 17434 | -0.959 | -0.3626 | No | ||
| 81 | EPHA7 | 17870 | -1.036 | -0.3749 | No | ||
| 82 | NFAM1 | 17899 | -1.042 | -0.3685 | No | ||
| 83 | HS6ST3 | 18218 | -1.096 | -0.3751 | No | ||
| 84 | VCL | 18239 | -1.100 | -0.3679 | No | ||
| 85 | VSNL1 | 18433 | -1.141 | -0.3684 | No | ||
| 86 | EIF4G1 | 18692 | -1.193 | -0.3714 | No | ||
| 87 | KCNN3 | 19213 | -1.314 | -0.3856 | No | ||
| 88 | ACCN4 | 19617 | -1.431 | -0.3935 | No | ||
| 89 | UBE4B | 20303 | -1.700 | -0.4124 | Yes | ||
| 90 | TRIB2 | 20441 | -1.762 | -0.4057 | Yes | ||
| 91 | NCOA5 | 20457 | -1.770 | -0.3934 | Yes | ||
| 92 | TM9SF1 | 20470 | -1.777 | -0.3809 | Yes | ||
| 93 | MNT | 20472 | -1.778 | -0.3679 | Yes | ||
| 94 | TMEM23 | 20554 | -1.824 | -0.3583 | Yes | ||
| 95 | LTA | 20785 | -1.958 | -0.3544 | Yes | ||
| 96 | MAML1 | 21132 | -2.222 | -0.3539 | Yes | ||
| 97 | UBTF | 21212 | -2.303 | -0.3407 | Yes | ||
| 98 | SHOX2 | 21345 | -2.479 | -0.3285 | Yes | ||
| 99 | ARF3 | 21349 | -2.483 | -0.3104 | Yes | ||
| 100 | SP6 | 21482 | -2.751 | -0.2963 | Yes | ||
| 101 | ZBTB9 | 21520 | -2.835 | -0.2771 | Yes | ||
| 102 | IRF1 | 21636 | -3.191 | -0.2590 | Yes | ||
| 103 | ZHX2 | 21654 | -3.267 | -0.2358 | Yes | ||
| 104 | RGS3 | 21752 | -3.840 | -0.2120 | Yes | ||
| 105 | PCBP4 | 21856 | -4.849 | -0.1811 | Yes | ||
| 106 | NR4A1 | 21923 | -6.633 | -0.1355 | Yes | ||
| 107 | EGR1 | 21942 | -8.847 | -0.0713 | Yes | ||
| 108 | EGR3 | 21943 | -9.744 | 0.0002 | Yes |