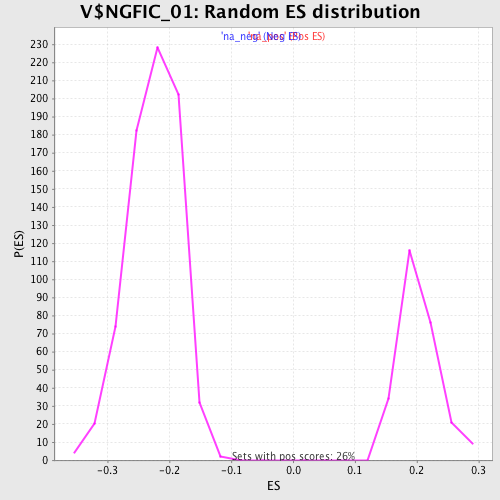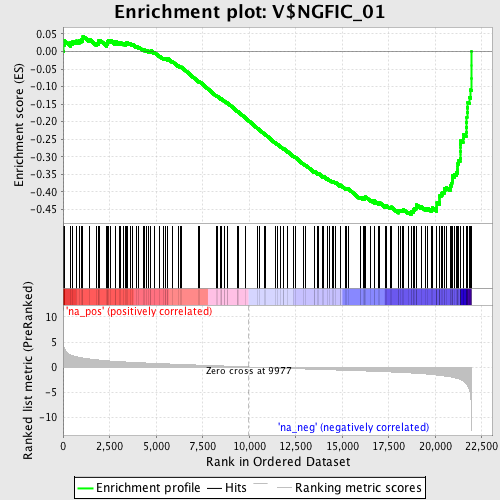
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NGFIC_01 |
| Enrichment Score (ES) | -0.46439525 |
| Normalized Enrichment Score (NES) | -2.0621843 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.027211E-4 |
| FWER p-Value | 0.0010 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ORC4L | 29 | 4.329 | 0.0167 | No | ||
| 2 | MATR3 | 55 | 3.775 | 0.0313 | No | ||
| 3 | BMPR2 | 409 | 2.410 | 0.0251 | No | ||
| 4 | NLK | 525 | 2.243 | 0.0292 | No | ||
| 5 | SFRS10 | 698 | 2.057 | 0.0299 | No | ||
| 6 | GABARAPL2 | 857 | 1.913 | 0.0306 | No | ||
| 7 | HNRPR | 963 | 1.844 | 0.0335 | No | ||
| 8 | ARHGAP12 | 1064 | 1.777 | 0.0363 | No | ||
| 9 | BCL11B | 1065 | 1.776 | 0.0437 | No | ||
| 10 | CENTG2 | 1403 | 1.602 | 0.0349 | No | ||
| 11 | LUC7L2 | 1778 | 1.447 | 0.0238 | No | ||
| 12 | SPG3A | 1892 | 1.397 | 0.0244 | No | ||
| 13 | FUS | 1896 | 1.396 | 0.0301 | No | ||
| 14 | DCX | 1980 | 1.366 | 0.0320 | No | ||
| 15 | UCHL3 | 2352 | 1.240 | 0.0201 | No | ||
| 16 | CLTC | 2387 | 1.228 | 0.0237 | No | ||
| 17 | SH3KBP1 | 2392 | 1.226 | 0.0286 | No | ||
| 18 | ETF1 | 2432 | 1.215 | 0.0319 | No | ||
| 19 | PCTK2 | 2547 | 1.187 | 0.0316 | No | ||
| 20 | NUDT11 | 2799 | 1.127 | 0.0247 | No | ||
| 21 | PABPC1 | 2803 | 1.126 | 0.0293 | No | ||
| 22 | FBXL11 | 3036 | 1.074 | 0.0231 | No | ||
| 23 | KLF3 | 3097 | 1.061 | 0.0248 | No | ||
| 24 | SYNCRIP | 3238 | 1.028 | 0.0227 | No | ||
| 25 | PPP1R12A | 3345 | 1.003 | 0.0220 | No | ||
| 26 | RPS6KA3 | 3383 | 0.995 | 0.0244 | No | ||
| 27 | COL27A1 | 3469 | 0.976 | 0.0246 | No | ||
| 28 | SNAP25 | 3593 | 0.949 | 0.0229 | No | ||
| 29 | RALGPS2 | 3719 | 0.926 | 0.0210 | No | ||
| 30 | KCNS3 | 3945 | 0.881 | 0.0144 | No | ||
| 31 | STC2 | 4062 | 0.861 | 0.0126 | No | ||
| 32 | NTRK3 | 4304 | 0.821 | 0.0050 | No | ||
| 33 | LRRC4 | 4387 | 0.805 | 0.0046 | No | ||
| 34 | CACNA1E | 4498 | 0.787 | 0.0028 | No | ||
| 35 | CGGBP1 | 4612 | 0.770 | 0.0008 | No | ||
| 36 | HNRPDL | 4715 | 0.753 | -0.0007 | No | ||
| 37 | KCNH3 | 4721 | 0.753 | 0.0022 | No | ||
| 38 | CNOT6L | 4899 | 0.726 | -0.0029 | No | ||
| 39 | MYB | 5181 | 0.683 | -0.0130 | No | ||
| 40 | VGF | 5406 | 0.648 | -0.0206 | No | ||
| 41 | EIF5A | 5477 | 0.638 | -0.0211 | No | ||
| 42 | NUDT10 | 5482 | 0.637 | -0.0186 | No | ||
| 43 | MRPL14 | 5628 | 0.618 | -0.0227 | No | ||
| 44 | MSI1 | 5630 | 0.618 | -0.0202 | No | ||
| 45 | SEP15 | 5887 | 0.576 | -0.0295 | No | ||
| 46 | ICAM5 | 6182 | 0.531 | -0.0408 | No | ||
| 47 | KCNJ1 | 6306 | 0.511 | -0.0443 | No | ||
| 48 | AXUD1 | 6350 | 0.506 | -0.0442 | No | ||
| 49 | KCNS2 | 7288 | 0.381 | -0.0857 | No | ||
| 50 | PITX2 | 7310 | 0.378 | -0.0850 | No | ||
| 51 | HAS1 | 8251 | 0.254 | -0.1271 | No | ||
| 52 | ADSS | 8299 | 0.247 | -0.1283 | No | ||
| 53 | KCNN2 | 8469 | 0.222 | -0.1351 | No | ||
| 54 | CNNM4 | 8492 | 0.218 | -0.1352 | No | ||
| 55 | WNT1 | 8503 | 0.217 | -0.1348 | No | ||
| 56 | KCNQ5 | 8660 | 0.195 | -0.1411 | No | ||
| 57 | ATP5G1 | 8683 | 0.192 | -0.1413 | No | ||
| 58 | NRD1 | 8812 | 0.174 | -0.1465 | No | ||
| 59 | ATP1B2 | 8845 | 0.169 | -0.1472 | No | ||
| 60 | FAF1 | 9349 | 0.098 | -0.1699 | No | ||
| 61 | ZNHIT1 | 9389 | 0.090 | -0.1713 | No | ||
| 62 | FEV | 9433 | 0.084 | -0.1730 | No | ||
| 63 | SH3GL3 | 9799 | 0.027 | -0.1896 | No | ||
| 64 | AP3S1 | 10442 | -0.069 | -0.2188 | No | ||
| 65 | LRFN5 | 10538 | -0.084 | -0.2228 | No | ||
| 66 | AAMP | 10798 | -0.122 | -0.2342 | No | ||
| 67 | BCL6 | 10847 | -0.131 | -0.2359 | No | ||
| 68 | EGLN2 | 11397 | -0.205 | -0.2602 | No | ||
| 69 | TBN | 11503 | -0.217 | -0.2641 | No | ||
| 70 | OTP | 11653 | -0.234 | -0.2700 | No | ||
| 71 | HMGN2 | 11848 | -0.257 | -0.2778 | No | ||
| 72 | NFYC | 11857 | -0.258 | -0.2771 | No | ||
| 73 | HYAL2 | 12082 | -0.281 | -0.2863 | No | ||
| 74 | PRPF3 | 12389 | -0.319 | -0.2990 | No | ||
| 75 | GLRA3 | 12485 | -0.329 | -0.3020 | No | ||
| 76 | BCL6B | 12936 | -0.383 | -0.3210 | No | ||
| 77 | KCNB1 | 13032 | -0.395 | -0.3238 | No | ||
| 78 | KCNA1 | 13486 | -0.449 | -0.3427 | No | ||
| 79 | CDH2 | 13529 | -0.455 | -0.3427 | No | ||
| 80 | FKBP2 | 13692 | -0.473 | -0.3482 | No | ||
| 81 | ACE | 13747 | -0.479 | -0.3487 | No | ||
| 82 | HOXA2 | 13955 | -0.504 | -0.3561 | No | ||
| 83 | HOXA7 | 14011 | -0.510 | -0.3565 | No | ||
| 84 | TNFRSF12A | 14180 | -0.530 | -0.3620 | No | ||
| 85 | VAX1 | 14334 | -0.549 | -0.3667 | No | ||
| 86 | REL | 14487 | -0.568 | -0.3713 | No | ||
| 87 | RFX4 | 14543 | -0.575 | -0.3715 | No | ||
| 88 | CDKN2C | 14633 | -0.587 | -0.3731 | No | ||
| 89 | SLC22A17 | 14877 | -0.615 | -0.3817 | No | ||
| 90 | JPH1 | 14904 | -0.619 | -0.3803 | No | ||
| 91 | SUMO2 | 15193 | -0.651 | -0.3908 | No | ||
| 92 | AMOT | 15220 | -0.654 | -0.3893 | No | ||
| 93 | SREBF2 | 15318 | -0.668 | -0.3909 | No | ||
| 94 | ACTN1 | 15953 | -0.750 | -0.4169 | No | ||
| 95 | PLOD3 | 15996 | -0.757 | -0.4157 | No | ||
| 96 | CRY2 | 16155 | -0.778 | -0.4197 | No | ||
| 97 | OTX1 | 16181 | -0.782 | -0.4176 | No | ||
| 98 | RAB2 | 16207 | -0.785 | -0.4155 | No | ||
| 99 | DVL2 | 16252 | -0.790 | -0.4142 | No | ||
| 100 | ASB7 | 16536 | -0.827 | -0.4238 | No | ||
| 101 | EFEMP2 | 16720 | -0.851 | -0.4286 | No | ||
| 102 | KCNIP2 | 16725 | -0.852 | -0.4252 | No | ||
| 103 | CENTG3 | 16958 | -0.885 | -0.4322 | No | ||
| 104 | PRR3 | 17015 | -0.893 | -0.4311 | No | ||
| 105 | HRK | 17305 | -0.939 | -0.4404 | No | ||
| 106 | RHOB | 17370 | -0.948 | -0.4394 | No | ||
| 107 | RELA | 17566 | -0.984 | -0.4442 | No | ||
| 108 | IHPK2 | 17628 | -0.995 | -0.4429 | No | ||
| 109 | ELAVL3 | 18006 | -1.060 | -0.4558 | No | ||
| 110 | POU3F1 | 18014 | -1.061 | -0.4517 | No | ||
| 111 | PDGFB | 18137 | -1.080 | -0.4528 | No | ||
| 112 | SMYD5 | 18225 | -1.098 | -0.4522 | No | ||
| 113 | UNC84B | 18292 | -1.111 | -0.4506 | No | ||
| 114 | PHF15 | 18556 | -1.165 | -0.4578 | No | ||
| 115 | NTN1 | 18700 | -1.194 | -0.4594 | Yes | ||
| 116 | SERTAD1 | 18702 | -1.195 | -0.4545 | Yes | ||
| 117 | JMJD1B | 18811 | -1.220 | -0.4544 | Yes | ||
| 118 | CACNA1A | 18814 | -1.221 | -0.4494 | Yes | ||
| 119 | GRB2 | 18879 | -1.237 | -0.4471 | Yes | ||
| 120 | TPM4 | 18962 | -1.254 | -0.4457 | Yes | ||
| 121 | GIT1 | 18987 | -1.260 | -0.4415 | Yes | ||
| 122 | HDAC9 | 19004 | -1.263 | -0.4370 | Yes | ||
| 123 | RBBP6 | 19244 | -1.322 | -0.4425 | Yes | ||
| 124 | APLP2 | 19476 | -1.388 | -0.4473 | Yes | ||
| 125 | GUCY1A2 | 19585 | -1.420 | -0.4463 | Yes | ||
| 126 | APP | 19814 | -1.503 | -0.4505 | Yes | ||
| 127 | PCDH17 | 19838 | -1.509 | -0.4453 | Yes | ||
| 128 | AP1G1 | 20050 | -1.590 | -0.4483 | Yes | ||
| 129 | KCND2 | 20081 | -1.602 | -0.4430 | Yes | ||
| 130 | PCSK2 | 20085 | -1.604 | -0.4365 | Yes | ||
| 131 | SEC14L1 | 20086 | -1.604 | -0.4298 | Yes | ||
| 132 | GRIN1 | 20215 | -1.658 | -0.4288 | Yes | ||
| 133 | MEF2C | 20238 | -1.670 | -0.4228 | Yes | ||
| 134 | ERF | 20240 | -1.671 | -0.4159 | Yes | ||
| 135 | RRBP1 | 20245 | -1.673 | -0.4091 | Yes | ||
| 136 | CLSTN1 | 20310 | -1.704 | -0.4049 | Yes | ||
| 137 | CX3CL1 | 20401 | -1.741 | -0.4018 | Yes | ||
| 138 | MNT | 20472 | -1.778 | -0.3976 | Yes | ||
| 139 | MYST2 | 20483 | -1.782 | -0.3906 | Yes | ||
| 140 | TLE3 | 20574 | -1.836 | -0.3871 | Yes | ||
| 141 | SAP130 | 20796 | -1.967 | -0.3891 | Yes | ||
| 142 | BAHD1 | 20822 | -1.978 | -0.3820 | Yes | ||
| 143 | SIN3A | 20857 | -2.001 | -0.3752 | Yes | ||
| 144 | KCNC1 | 20897 | -2.031 | -0.3685 | Yes | ||
| 145 | GNAI2 | 20916 | -2.045 | -0.3608 | Yes | ||
| 146 | LRP10 | 20931 | -2.056 | -0.3529 | Yes | ||
| 147 | NFE2L1 | 21046 | -2.149 | -0.3491 | Yes | ||
| 148 | SH3BP1 | 21143 | -2.229 | -0.3443 | Yes | ||
| 149 | MAN2A2 | 21198 | -2.293 | -0.3372 | Yes | ||
| 150 | MADD | 21207 | -2.298 | -0.3280 | Yes | ||
| 151 | UBTF | 21212 | -2.303 | -0.3186 | Yes | ||
| 152 | ERBB3 | 21264 | -2.361 | -0.3110 | Yes | ||
| 153 | BHLHB2 | 21329 | -2.459 | -0.3037 | Yes | ||
| 154 | MAP3K4 | 21340 | -2.470 | -0.2939 | Yes | ||
| 155 | RASGEF1A | 21341 | -2.470 | -0.2836 | Yes | ||
| 156 | ARF3 | 21349 | -2.483 | -0.2736 | Yes | ||
| 157 | SLC25A5 | 21353 | -2.489 | -0.2633 | Yes | ||
| 158 | ZFPM1 | 21370 | -2.515 | -0.2536 | Yes | ||
| 159 | BDNF | 21491 | -2.758 | -0.2476 | Yes | ||
| 160 | SMAD3 | 21501 | -2.785 | -0.2364 | Yes | ||
| 161 | ZHX2 | 21654 | -3.267 | -0.2297 | Yes | ||
| 162 | NRGN | 21664 | -3.322 | -0.2163 | Yes | ||
| 163 | TRIB1 | 21684 | -3.431 | -0.2029 | Yes | ||
| 164 | TLE4 | 21695 | -3.506 | -0.1887 | Yes | ||
| 165 | CORO1C | 21703 | -3.548 | -0.1743 | Yes | ||
| 166 | PDCD1 | 21717 | -3.625 | -0.1597 | Yes | ||
| 167 | RELB | 21749 | -3.826 | -0.1452 | Yes | ||
| 168 | LASP1 | 21823 | -4.444 | -0.1300 | Yes | ||
| 169 | PJA1 | 21889 | -5.620 | -0.1096 | Yes | ||
| 170 | LEF1 | 21939 | -8.298 | -0.0772 | Yes | ||
| 171 | EGR1 | 21942 | -8.847 | -0.0404 | Yes | ||
| 172 | EGR3 | 21943 | -9.744 | 0.0002 | Yes |