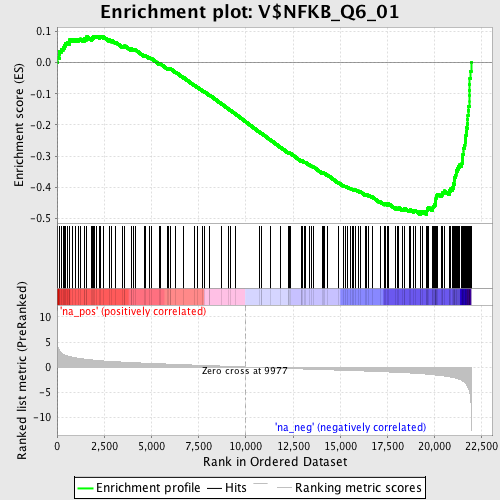
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NFKB_Q6_01 |
| Enrichment Score (ES) | -0.48646706 |
| Normalized Enrichment Score (NES) | -2.1401322 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL2RA | 15 | 4.747 | 0.0170 | No | ||
| 2 | CHD6 | 116 | 3.310 | 0.0247 | No | ||
| 3 | CREB1 | 126 | 3.257 | 0.0363 | No | ||
| 4 | IGSF4 | 212 | 2.865 | 0.0431 | No | ||
| 5 | ASH1L | 314 | 2.583 | 0.0481 | No | ||
| 6 | ANKHD1 | 395 | 2.431 | 0.0534 | No | ||
| 7 | REV3L | 423 | 2.391 | 0.0611 | No | ||
| 8 | NLK | 525 | 2.243 | 0.0648 | No | ||
| 9 | RBMS1 | 666 | 2.092 | 0.0661 | No | ||
| 10 | BAZ2B | 677 | 2.083 | 0.0734 | No | ||
| 11 | FGF17 | 816 | 1.949 | 0.0743 | No | ||
| 12 | VCAM1 | 984 | 1.831 | 0.0734 | No | ||
| 13 | PTGES | 1114 | 1.751 | 0.0740 | No | ||
| 14 | GNB1 | 1222 | 1.696 | 0.0754 | No | ||
| 15 | GPHN | 1432 | 1.593 | 0.0717 | No | ||
| 16 | CHD4 | 1458 | 1.578 | 0.0764 | No | ||
| 17 | ATP1B1 | 1557 | 1.539 | 0.0776 | No | ||
| 18 | BMF | 1578 | 1.530 | 0.0824 | No | ||
| 19 | NEK8 | 1831 | 1.422 | 0.0761 | No | ||
| 20 | PTPRJ | 1863 | 1.408 | 0.0799 | No | ||
| 21 | NR3C1 | 1927 | 1.383 | 0.0822 | No | ||
| 22 | NFAT5 | 1993 | 1.363 | 0.0843 | No | ||
| 23 | MITF | 2097 | 1.330 | 0.0845 | No | ||
| 24 | BMP2K | 2252 | 1.276 | 0.0821 | No | ||
| 25 | SOCS2 | 2303 | 1.259 | 0.0845 | No | ||
| 26 | SOX5 | 2456 | 1.209 | 0.0820 | No | ||
| 27 | DDR1 | 2758 | 1.137 | 0.0724 | No | ||
| 28 | ALG6 | 2907 | 1.105 | 0.0698 | No | ||
| 29 | TOP1 | 3099 | 1.061 | 0.0649 | No | ||
| 30 | COL27A1 | 3469 | 0.976 | 0.0516 | No | ||
| 31 | RAP2C | 3553 | 0.957 | 0.0514 | No | ||
| 32 | SNAP25 | 3593 | 0.949 | 0.0531 | No | ||
| 33 | ERN1 | 3917 | 0.886 | 0.0416 | No | ||
| 34 | FBXL12 | 3928 | 0.885 | 0.0444 | No | ||
| 35 | SLC6A12 | 4059 | 0.862 | 0.0416 | No | ||
| 36 | AP1S2 | 4155 | 0.845 | 0.0404 | No | ||
| 37 | CCL5 | 4636 | 0.767 | 0.0212 | No | ||
| 38 | KRTAP13-1 | 4694 | 0.757 | 0.0214 | No | ||
| 39 | RBPSUH | 4891 | 0.726 | 0.0151 | No | ||
| 40 | UBE2D3 | 4979 | 0.711 | 0.0138 | No | ||
| 41 | RIN2 | 5426 | 0.645 | -0.0043 | No | ||
| 42 | EIF5A | 5477 | 0.638 | -0.0043 | No | ||
| 43 | EP300 | 5873 | 0.578 | -0.0202 | No | ||
| 44 | PAPPA | 5897 | 0.575 | -0.0192 | No | ||
| 45 | ZBTB11 | 5985 | 0.561 | -0.0211 | No | ||
| 46 | FGF1 | 5996 | 0.558 | -0.0195 | No | ||
| 47 | MOBKL2C | 6294 | 0.513 | -0.0312 | No | ||
| 48 | C1QL1 | 6719 | 0.457 | -0.0490 | No | ||
| 49 | EDG5 | 7274 | 0.384 | -0.0730 | No | ||
| 50 | FUT7 | 7456 | 0.359 | -0.0799 | No | ||
| 51 | HTR3B | 7717 | 0.325 | -0.0907 | No | ||
| 52 | CSF3 | 7806 | 0.314 | -0.0936 | No | ||
| 53 | KLK9 | 8062 | 0.279 | -0.1042 | No | ||
| 54 | IFNB1 | 8083 | 0.276 | -0.1041 | No | ||
| 55 | IL23A | 8716 | 0.188 | -0.1324 | No | ||
| 56 | ILK | 9095 | 0.134 | -0.1493 | No | ||
| 57 | UACA | 9191 | 0.121 | -0.1532 | No | ||
| 58 | ARHGAP8 | 9477 | 0.078 | -0.1660 | No | ||
| 59 | MSX1 | 10717 | -0.110 | -0.2225 | No | ||
| 60 | EGF | 10831 | -0.129 | -0.2272 | No | ||
| 61 | MSC | 10845 | -0.131 | -0.2273 | No | ||
| 62 | CLCN1 | 11298 | -0.194 | -0.2474 | No | ||
| 63 | TRIM47 | 11828 | -0.255 | -0.2707 | No | ||
| 64 | TSLP | 12250 | -0.301 | -0.2889 | No | ||
| 65 | MAG | 12268 | -0.303 | -0.2886 | No | ||
| 66 | GREM1 | 12314 | -0.311 | -0.2895 | No | ||
| 67 | CENTB5 | 12387 | -0.319 | -0.2916 | No | ||
| 68 | CNTNAP1 | 12935 | -0.383 | -0.3153 | No | ||
| 69 | BCL6B | 12936 | -0.383 | -0.3139 | No | ||
| 70 | IL12A | 12986 | -0.388 | -0.3147 | No | ||
| 71 | SMOC1 | 13129 | -0.405 | -0.3197 | No | ||
| 72 | ASCL3 | 13150 | -0.408 | -0.3191 | No | ||
| 73 | PFN1 | 13354 | -0.432 | -0.3268 | No | ||
| 74 | MAP3K11 | 13487 | -0.449 | -0.3312 | No | ||
| 75 | POU2F3 | 13591 | -0.459 | -0.3342 | No | ||
| 76 | PTHLH | 14038 | -0.513 | -0.3528 | No | ||
| 77 | DUSP22 | 14089 | -0.518 | -0.3532 | No | ||
| 78 | UPF2 | 14163 | -0.528 | -0.3546 | No | ||
| 79 | RND1 | 14328 | -0.548 | -0.3600 | No | ||
| 80 | GRIN2D | 14936 | -0.622 | -0.3856 | No | ||
| 81 | RASGRP4 | 15158 | -0.647 | -0.3934 | No | ||
| 82 | IL13 | 15257 | -0.658 | -0.3954 | No | ||
| 83 | SIRT2 | 15387 | -0.675 | -0.3988 | No | ||
| 84 | GATA4 | 15522 | -0.689 | -0.4024 | No | ||
| 85 | SMPD3 | 15637 | -0.706 | -0.4050 | No | ||
| 86 | BAI2 | 15719 | -0.717 | -0.4061 | No | ||
| 87 | MRPS6 | 15817 | -0.731 | -0.4078 | No | ||
| 88 | ACTN1 | 15953 | -0.750 | -0.4112 | No | ||
| 89 | TCF1 | 16059 | -0.765 | -0.4132 | No | ||
| 90 | AMOTL1 | 16365 | -0.806 | -0.4242 | No | ||
| 91 | NFKBIA | 16391 | -0.809 | -0.4224 | No | ||
| 92 | UBD | 16505 | -0.824 | -0.4245 | No | ||
| 93 | HSD3B7 | 16699 | -0.847 | -0.4302 | No | ||
| 94 | RALGDS | 17121 | -0.911 | -0.4461 | No | ||
| 95 | BIRC3 | 17339 | -0.944 | -0.4526 | No | ||
| 96 | WNT10A | 17395 | -0.953 | -0.4516 | No | ||
| 97 | ETV1 | 17507 | -0.973 | -0.4531 | No | ||
| 98 | MAML2 | 17538 | -0.979 | -0.4508 | No | ||
| 99 | TGIF | 17938 | -1.050 | -0.4652 | No | ||
| 100 | IL1RN | 18058 | -1.069 | -0.4667 | No | ||
| 101 | PHOX2B | 18114 | -1.078 | -0.4652 | No | ||
| 102 | UNC84B | 18292 | -1.111 | -0.4692 | No | ||
| 103 | DGKI | 18390 | -1.133 | -0.4695 | No | ||
| 104 | UBE2H | 18421 | -1.140 | -0.4666 | No | ||
| 105 | BLR1 | 18680 | -1.189 | -0.4741 | No | ||
| 106 | NTN1 | 18700 | -1.194 | -0.4705 | No | ||
| 107 | NFKBIB | 18902 | -1.241 | -0.4751 | No | ||
| 108 | CYLD | 18965 | -1.255 | -0.4733 | No | ||
| 109 | DCAMKL1 | 19253 | -1.325 | -0.4815 | Yes | ||
| 110 | RHOG | 19258 | -1.326 | -0.4768 | Yes | ||
| 111 | TNIP1 | 19385 | -1.362 | -0.4775 | Yes | ||
| 112 | MADCAM1 | 19568 | -1.416 | -0.4806 | Yes | ||
| 113 | RFX5 | 19571 | -1.417 | -0.4754 | Yes | ||
| 114 | TCEA2 | 19600 | -1.424 | -0.4714 | Yes | ||
| 115 | DAP3 | 19633 | -1.434 | -0.4676 | Yes | ||
| 116 | TP53 | 19687 | -1.455 | -0.4646 | Yes | ||
| 117 | IL7 | 19888 | -1.525 | -0.4681 | Yes | ||
| 118 | SEMA3B | 19915 | -1.535 | -0.4636 | Yes | ||
| 119 | ZNRF1 | 19955 | -1.549 | -0.4596 | Yes | ||
| 120 | EDN2 | 19996 | -1.569 | -0.4556 | Yes | ||
| 121 | HIVEP1 | 20031 | -1.583 | -0.4513 | Yes | ||
| 122 | LTB | 20040 | -1.586 | -0.4458 | Yes | ||
| 123 | GNG4 | 20044 | -1.587 | -0.4400 | Yes | ||
| 124 | ZIC4 | 20073 | -1.599 | -0.4354 | Yes | ||
| 125 | PCSK2 | 20085 | -1.604 | -0.4299 | Yes | ||
| 126 | TNFSF7 | 20122 | -1.618 | -0.4256 | Yes | ||
| 127 | EHF | 20162 | -1.636 | -0.4213 | Yes | ||
| 128 | SLAMF8 | 20358 | -1.724 | -0.4238 | Yes | ||
| 129 | TBX20 | 20399 | -1.741 | -0.4192 | Yes | ||
| 130 | TRIB2 | 20441 | -1.762 | -0.4145 | Yes | ||
| 131 | PRKCD | 20514 | -1.805 | -0.4111 | Yes | ||
| 132 | LTA | 20785 | -1.958 | -0.4163 | Yes | ||
| 133 | CD69 | 20802 | -1.970 | -0.4097 | Yes | ||
| 134 | SIN3A | 20857 | -2.001 | -0.4047 | Yes | ||
| 135 | SEC61A1 | 20950 | -2.069 | -0.4012 | Yes | ||
| 136 | TRAF4 | 21000 | -2.116 | -0.3956 | Yes | ||
| 137 | NR2F2 | 21012 | -2.123 | -0.3882 | Yes | ||
| 138 | EBI3 | 21047 | -2.149 | -0.3818 | Yes | ||
| 139 | IL6ST | 21050 | -2.151 | -0.3739 | Yes | ||
| 140 | USP52 | 21073 | -2.166 | -0.3669 | Yes | ||
| 141 | ICAM1 | 21114 | -2.204 | -0.3605 | Yes | ||
| 142 | UBE2I | 21139 | -2.226 | -0.3534 | Yes | ||
| 143 | MAP4K2 | 21147 | -2.232 | -0.3454 | Yes | ||
| 144 | NDRG2 | 21223 | -2.314 | -0.3402 | Yes | ||
| 145 | GRK5 | 21250 | -2.344 | -0.3327 | Yes | ||
| 146 | SHOX2 | 21345 | -2.479 | -0.3278 | Yes | ||
| 147 | RASSF2 | 21452 | -2.692 | -0.3227 | Yes | ||
| 148 | SP6 | 21482 | -2.751 | -0.3138 | Yes | ||
| 149 | BDNF | 21491 | -2.758 | -0.3039 | Yes | ||
| 150 | BATF | 21502 | -2.787 | -0.2940 | Yes | ||
| 151 | STAT6 | 21519 | -2.830 | -0.2842 | Yes | ||
| 152 | ZBTB9 | 21520 | -2.835 | -0.2737 | Yes | ||
| 153 | JAK3 | 21578 | -3.015 | -0.2651 | Yes | ||
| 154 | NFKB2 | 21615 | -3.137 | -0.2551 | Yes | ||
| 155 | ELMO1 | 21628 | -3.167 | -0.2439 | Yes | ||
| 156 | IER5 | 21661 | -3.308 | -0.2331 | Yes | ||
| 157 | MMP9 | 21679 | -3.409 | -0.2212 | Yes | ||
| 158 | ENO3 | 21711 | -3.589 | -0.2093 | Yes | ||
| 159 | XPO6 | 21734 | -3.713 | -0.1965 | Yes | ||
| 160 | RELB | 21749 | -3.826 | -0.1829 | Yes | ||
| 161 | ARHGEF2 | 21769 | -3.994 | -0.1689 | Yes | ||
| 162 | TNFRSF9 | 21782 | -4.110 | -0.1542 | Yes | ||
| 163 | GADD45B | 21806 | -4.294 | -0.1393 | Yes | ||
| 164 | IL4I1 | 21849 | -4.754 | -0.1236 | Yes | ||
| 165 | TNFRSF1B | 21851 | -4.763 | -0.1059 | Yes | ||
| 166 | PCBP4 | 21856 | -4.849 | -0.0881 | Yes | ||
| 167 | BCL3 | 21865 | -4.977 | -0.0700 | Yes | ||
| 168 | IL27RA | 21874 | -5.232 | -0.0509 | Yes | ||
| 169 | PPP1R13B | 21915 | -6.417 | -0.0289 | Yes | ||
| 170 | DUSP6 | 21938 | -8.151 | 0.0004 | Yes |
