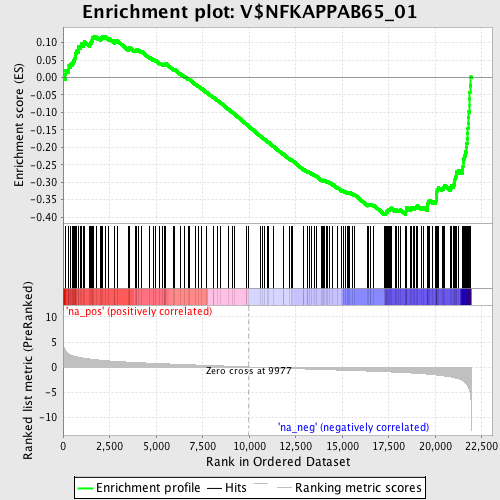
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NFKAPPAB65_01 |
| Enrichment Score (ES) | -0.39231744 |
| Normalized Enrichment Score (NES) | -1.7629728 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.016059194 |
| FWER p-Value | 0.207 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CHD6 | 116 | 3.310 | 0.0081 | No | ||
| 2 | CREB1 | 126 | 3.257 | 0.0208 | No | ||
| 3 | SLC12A2 | 297 | 2.620 | 0.0236 | No | ||
| 4 | ASH1L | 314 | 2.583 | 0.0333 | No | ||
| 5 | TCF8 | 411 | 2.409 | 0.0387 | No | ||
| 6 | NLK | 525 | 2.243 | 0.0425 | No | ||
| 7 | CDK6 | 570 | 2.184 | 0.0494 | No | ||
| 8 | YWHAZ | 622 | 2.135 | 0.0557 | No | ||
| 9 | RBMS1 | 666 | 2.092 | 0.0621 | No | ||
| 10 | BAZ2B | 677 | 2.083 | 0.0701 | No | ||
| 11 | TATDN1 | 730 | 2.027 | 0.0759 | No | ||
| 12 | CLOCK | 811 | 1.956 | 0.0802 | No | ||
| 13 | FGF17 | 816 | 1.949 | 0.0879 | No | ||
| 14 | PCDH10 | 951 | 1.852 | 0.0892 | No | ||
| 15 | HNRPR | 963 | 1.844 | 0.0961 | No | ||
| 16 | PTGES | 1114 | 1.751 | 0.0963 | No | ||
| 17 | SEC63 | 1139 | 1.737 | 0.1023 | No | ||
| 18 | TIAL1 | 1439 | 1.589 | 0.0950 | No | ||
| 19 | CHD4 | 1458 | 1.578 | 0.1005 | No | ||
| 20 | SIX4 | 1508 | 1.560 | 0.1046 | No | ||
| 21 | ATP1B1 | 1557 | 1.539 | 0.1086 | No | ||
| 22 | BMF | 1578 | 1.530 | 0.1139 | No | ||
| 23 | CD86 | 1658 | 1.497 | 0.1163 | No | ||
| 24 | ZDHHC8 | 1792 | 1.441 | 0.1160 | No | ||
| 25 | NFAT5 | 1993 | 1.363 | 0.1123 | No | ||
| 26 | WRN | 2060 | 1.342 | 0.1147 | No | ||
| 27 | MITF | 2097 | 1.330 | 0.1185 | No | ||
| 28 | BMP2K | 2252 | 1.276 | 0.1165 | No | ||
| 29 | SOX5 | 2456 | 1.209 | 0.1121 | No | ||
| 30 | DDR1 | 2758 | 1.137 | 0.1029 | No | ||
| 31 | LAMA1 | 2771 | 1.133 | 0.1069 | No | ||
| 32 | ALG6 | 2907 | 1.105 | 0.1052 | No | ||
| 33 | IER3 | 3521 | 0.965 | 0.0809 | No | ||
| 34 | RAP2C | 3553 | 0.957 | 0.0834 | No | ||
| 35 | CASKIN2 | 3572 | 0.954 | 0.0864 | No | ||
| 36 | TRPC4 | 3862 | 0.899 | 0.0768 | No | ||
| 37 | ERN1 | 3917 | 0.886 | 0.0779 | No | ||
| 38 | AKT1S1 | 3957 | 0.880 | 0.0796 | No | ||
| 39 | SLC6A12 | 4059 | 0.862 | 0.0785 | No | ||
| 40 | CLCN2 | 4195 | 0.840 | 0.0757 | No | ||
| 41 | CCL5 | 4636 | 0.767 | 0.0586 | No | ||
| 42 | IL1RAPL1 | 4833 | 0.734 | 0.0525 | No | ||
| 43 | UBE2D3 | 4979 | 0.711 | 0.0487 | No | ||
| 44 | NDUFB9 | 5203 | 0.679 | 0.0412 | No | ||
| 45 | DSC2 | 5357 | 0.656 | 0.0369 | No | ||
| 46 | RIN2 | 5426 | 0.645 | 0.0364 | No | ||
| 47 | FGF12 | 5464 | 0.640 | 0.0372 | No | ||
| 48 | EIF5A | 5477 | 0.638 | 0.0393 | No | ||
| 49 | MAPK6 | 5506 | 0.633 | 0.0406 | No | ||
| 50 | COL11A2 | 5940 | 0.568 | 0.0230 | No | ||
| 51 | FGF1 | 5996 | 0.558 | 0.0227 | No | ||
| 52 | MOBKL2C | 6294 | 0.513 | 0.0111 | No | ||
| 53 | TSNAXIP1 | 6539 | 0.482 | 0.0019 | No | ||
| 54 | C1QL1 | 6719 | 0.457 | -0.0045 | No | ||
| 55 | SLC16A6 | 6778 | 0.449 | -0.0054 | No | ||
| 56 | E2F3 | 7113 | 0.406 | -0.0191 | No | ||
| 57 | EDG5 | 7274 | 0.384 | -0.0249 | No | ||
| 58 | FUT7 | 7456 | 0.359 | -0.0317 | No | ||
| 59 | HTR3B | 7717 | 0.325 | -0.0424 | No | ||
| 60 | KLK9 | 8062 | 0.279 | -0.0570 | No | ||
| 61 | IFNB1 | 8083 | 0.276 | -0.0568 | No | ||
| 62 | CTDSP1 | 8301 | 0.247 | -0.0658 | No | ||
| 63 | KCNN2 | 8469 | 0.222 | -0.0726 | No | ||
| 64 | NXPH4 | 8901 | 0.162 | -0.0917 | No | ||
| 65 | ILK | 9095 | 0.134 | -0.1001 | No | ||
| 66 | UACA | 9191 | 0.121 | -0.1039 | No | ||
| 67 | ADCK4 | 9852 | 0.019 | -0.1342 | No | ||
| 68 | SIX5 | 9872 | 0.016 | -0.1350 | No | ||
| 69 | EIF4A2 | 9987 | -0.002 | -0.1402 | No | ||
| 70 | TBC1D17 | 10605 | -0.095 | -0.1682 | No | ||
| 71 | MSX1 | 10717 | -0.110 | -0.1728 | No | ||
| 72 | MSC | 10845 | -0.131 | -0.1781 | No | ||
| 73 | WNT10B | 10979 | -0.152 | -0.1836 | No | ||
| 74 | FLOT1 | 11042 | -0.161 | -0.1858 | No | ||
| 75 | CLCN1 | 11298 | -0.194 | -0.1968 | No | ||
| 76 | ACTN3 | 11825 | -0.254 | -0.2199 | No | ||
| 77 | TRIM47 | 11828 | -0.255 | -0.2190 | No | ||
| 78 | COL16A1 | 12170 | -0.292 | -0.2335 | No | ||
| 79 | TSLP | 12250 | -0.301 | -0.2359 | No | ||
| 80 | GREM1 | 12314 | -0.311 | -0.2375 | No | ||
| 81 | PURG | 12929 | -0.383 | -0.2642 | No | ||
| 82 | BCL6B | 12936 | -0.383 | -0.2629 | No | ||
| 83 | SMOC1 | 13129 | -0.405 | -0.2701 | No | ||
| 84 | ASCL3 | 13150 | -0.408 | -0.2693 | No | ||
| 85 | MAP3K8 | 13234 | -0.417 | -0.2715 | No | ||
| 86 | PFN1 | 13354 | -0.432 | -0.2752 | No | ||
| 87 | MAP3K11 | 13487 | -0.449 | -0.2794 | No | ||
| 88 | POU2F3 | 13591 | -0.459 | -0.2823 | No | ||
| 89 | WNT4 | 13890 | -0.497 | -0.2940 | No | ||
| 90 | CLDN5 | 13961 | -0.504 | -0.2952 | No | ||
| 91 | GPM6A | 13971 | -0.505 | -0.2935 | No | ||
| 92 | PTHLH | 14038 | -0.513 | -0.2945 | No | ||
| 93 | UPF2 | 14163 | -0.528 | -0.2981 | No | ||
| 94 | AGC1 | 14232 | -0.536 | -0.2990 | No | ||
| 95 | RND1 | 14328 | -0.548 | -0.3012 | No | ||
| 96 | REL | 14487 | -0.568 | -0.3061 | No | ||
| 97 | RPS19 | 14743 | -0.600 | -0.3154 | No | ||
| 98 | BLCAP | 14970 | -0.625 | -0.3233 | No | ||
| 99 | YWHAQ | 15062 | -0.634 | -0.3249 | No | ||
| 100 | RASGRP4 | 15158 | -0.647 | -0.3266 | No | ||
| 101 | IL13 | 15257 | -0.658 | -0.3285 | No | ||
| 102 | CACNG3 | 15337 | -0.669 | -0.3294 | No | ||
| 103 | SIRT2 | 15387 | -0.675 | -0.3289 | No | ||
| 104 | GATA4 | 15522 | -0.689 | -0.3323 | No | ||
| 105 | SMPD3 | 15637 | -0.706 | -0.3346 | No | ||
| 106 | AMOTL1 | 16365 | -0.806 | -0.3648 | No | ||
| 107 | NFKBIA | 16391 | -0.809 | -0.3627 | No | ||
| 108 | UBD | 16505 | -0.824 | -0.3645 | No | ||
| 109 | CXCL11 | 16542 | -0.828 | -0.3628 | No | ||
| 110 | HSD3B7 | 16699 | -0.847 | -0.3666 | No | ||
| 111 | PLXNB1 | 17260 | -0.931 | -0.3885 | Yes | ||
| 112 | BIRC3 | 17339 | -0.944 | -0.3883 | Yes | ||
| 113 | RRAS | 17359 | -0.947 | -0.3853 | Yes | ||
| 114 | WNT10A | 17395 | -0.953 | -0.3831 | Yes | ||
| 115 | SOX10 | 17450 | -0.962 | -0.3817 | Yes | ||
| 116 | SDC4 | 17459 | -0.965 | -0.3781 | Yes | ||
| 117 | MAML2 | 17538 | -0.979 | -0.3778 | Yes | ||
| 118 | CUEDC1 | 17576 | -0.986 | -0.3755 | Yes | ||
| 119 | SOX3 | 17626 | -0.995 | -0.3737 | Yes | ||
| 120 | LRCH1 | 17840 | -1.029 | -0.3793 | Yes | ||
| 121 | ABI3 | 17927 | -1.047 | -0.3790 | Yes | ||
| 122 | DOCK4 | 18046 | -1.067 | -0.3801 | Yes | ||
| 123 | RPS6KA4 | 18117 | -1.078 | -0.3790 | Yes | ||
| 124 | GNGT2 | 18408 | -1.137 | -0.3877 | Yes | ||
| 125 | UBE2H | 18421 | -1.140 | -0.3837 | Yes | ||
| 126 | TLX3 | 18428 | -1.141 | -0.3793 | Yes | ||
| 127 | TLX1 | 18435 | -1.142 | -0.3750 | Yes | ||
| 128 | FKHL18 | 18450 | -1.144 | -0.3710 | Yes | ||
| 129 | BLR1 | 18680 | -1.189 | -0.3767 | Yes | ||
| 130 | EIF4G1 | 18692 | -1.193 | -0.3724 | Yes | ||
| 131 | PCDH12 | 18806 | -1.218 | -0.3726 | Yes | ||
| 132 | NFKBIB | 18902 | -1.241 | -0.3720 | Yes | ||
| 133 | CYLD | 18965 | -1.255 | -0.3698 | Yes | ||
| 134 | CSF1R | 19015 | -1.265 | -0.3669 | Yes | ||
| 135 | DCAMKL1 | 19253 | -1.325 | -0.3724 | Yes | ||
| 136 | TNIP1 | 19385 | -1.362 | -0.3729 | Yes | ||
| 137 | MADCAM1 | 19568 | -1.416 | -0.3756 | Yes | ||
| 138 | RFX5 | 19571 | -1.417 | -0.3699 | Yes | ||
| 139 | RANBP10 | 19574 | -1.417 | -0.3643 | Yes | ||
| 140 | TCEA2 | 19600 | -1.424 | -0.3597 | Yes | ||
| 141 | DAP3 | 19633 | -1.434 | -0.3553 | Yes | ||
| 142 | TP53 | 19687 | -1.455 | -0.3519 | Yes | ||
| 143 | CXCL10 | 19873 | -1.519 | -0.3542 | Yes | ||
| 144 | HIVEP1 | 20031 | -1.583 | -0.3551 | Yes | ||
| 145 | LTB | 20040 | -1.586 | -0.3490 | Yes | ||
| 146 | GNG4 | 20044 | -1.587 | -0.3427 | Yes | ||
| 147 | ITPKC | 20064 | -1.594 | -0.3371 | Yes | ||
| 148 | ZIC4 | 20073 | -1.599 | -0.3310 | Yes | ||
| 149 | PCSK2 | 20085 | -1.604 | -0.3251 | Yes | ||
| 150 | TNFSF7 | 20122 | -1.618 | -0.3202 | Yes | ||
| 151 | EHF | 20162 | -1.636 | -0.3153 | Yes | ||
| 152 | SLAMF8 | 20358 | -1.724 | -0.3173 | Yes | ||
| 153 | TRIB2 | 20441 | -1.762 | -0.3140 | Yes | ||
| 154 | MYST2 | 20483 | -1.782 | -0.3086 | Yes | ||
| 155 | CD69 | 20802 | -1.970 | -0.3153 | Yes | ||
| 156 | SIN3A | 20857 | -2.001 | -0.3097 | Yes | ||
| 157 | TRAF4 | 21000 | -2.116 | -0.3076 | Yes | ||
| 158 | NR2F2 | 21012 | -2.123 | -0.2995 | Yes | ||
| 159 | IL6ST | 21050 | -2.151 | -0.2925 | Yes | ||
| 160 | USP52 | 21073 | -2.166 | -0.2848 | Yes | ||
| 161 | ICAM1 | 21114 | -2.204 | -0.2777 | Yes | ||
| 162 | UBE2I | 21139 | -2.226 | -0.2698 | Yes | ||
| 163 | GRK5 | 21250 | -2.344 | -0.2654 | Yes | ||
| 164 | RASSF2 | 21452 | -2.692 | -0.2637 | Yes | ||
| 165 | SP6 | 21482 | -2.751 | -0.2539 | Yes | ||
| 166 | BDNF | 21491 | -2.758 | -0.2431 | Yes | ||
| 167 | STAT6 | 21519 | -2.830 | -0.2329 | Yes | ||
| 168 | JAK3 | 21578 | -3.015 | -0.2234 | Yes | ||
| 169 | NFKB2 | 21615 | -3.137 | -0.2124 | Yes | ||
| 170 | IER5 | 21661 | -3.308 | -0.2010 | Yes | ||
| 171 | MMP9 | 21679 | -3.409 | -0.1880 | Yes | ||
| 172 | ENO3 | 21711 | -3.589 | -0.1749 | Yes | ||
| 173 | ZMYND15 | 21743 | -3.778 | -0.1611 | Yes | ||
| 174 | RELB | 21749 | -3.826 | -0.1458 | Yes | ||
| 175 | ARHGEF2 | 21769 | -3.994 | -0.1305 | Yes | ||
| 176 | TNFRSF9 | 21782 | -4.110 | -0.1145 | Yes | ||
| 177 | GADD45B | 21806 | -4.294 | -0.0982 | Yes | ||
| 178 | TNFRSF1B | 21851 | -4.763 | -0.0809 | Yes | ||
| 179 | CXCL16 | 21853 | -4.800 | -0.0615 | Yes | ||
| 180 | PCBP4 | 21856 | -4.849 | -0.0420 | Yes | ||
| 181 | BCL3 | 21865 | -4.977 | -0.0222 | Yes | ||
| 182 | PPP1R13B | 21915 | -6.417 | 0.0015 | Yes |
