Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$FAC1_01 |
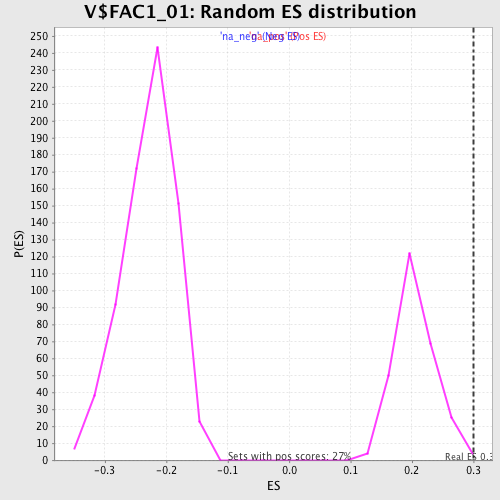
| Enrichment Score (ES) | 0.29970112 |
| Normalized Enrichment Score (NES) | 1.4584895 |
| Nominal p-value | 0.00729927 |
| FDR q-value | 0.19657263 |
| FWER p-Value | 0.866 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PDGFRB | 6 | 5.817 | 0.0316 | Yes | ||
| 2 | SFRS12 | 33 | 4.169 | 0.0532 | Yes | ||
| 3 | CNOT4 | 51 | 3.789 | 0.0732 | Yes | ||
| 4 | CHD6 | 116 | 3.310 | 0.0884 | Yes | ||
| 5 | TCF7L2 | 168 | 3.009 | 0.1026 | Yes | ||
| 6 | FBLN2 | 227 | 2.827 | 0.1154 | Yes | ||
| 7 | USP34 | 241 | 2.782 | 0.1301 | Yes | ||
| 8 | FBXL20 | 393 | 2.432 | 0.1365 | Yes | ||
| 9 | H3F3A | 403 | 2.420 | 0.1493 | Yes | ||
| 10 | BRCA1 | 426 | 2.388 | 0.1614 | Yes | ||
| 11 | RUNX1 | 523 | 2.244 | 0.1693 | Yes | ||
| 12 | RBMS1 | 666 | 2.092 | 0.1742 | Yes | ||
| 13 | JARID2 | 776 | 1.986 | 0.1801 | Yes | ||
| 14 | GABARAPL2 | 857 | 1.913 | 0.1869 | Yes | ||
| 15 | GNAS | 956 | 1.850 | 0.1925 | Yes | ||
| 16 | CTLA4 | 1130 | 1.742 | 0.1942 | Yes | ||
| 17 | CHM | 1194 | 1.711 | 0.2006 | Yes | ||
| 18 | KCNMA1 | 1226 | 1.694 | 0.2085 | Yes | ||
| 19 | FOXP1 | 1454 | 1.579 | 0.2067 | Yes | ||
| 20 | SIX4 | 1508 | 1.560 | 0.2129 | Yes | ||
| 21 | HFE2 | 1516 | 1.556 | 0.2211 | Yes | ||
| 22 | RAB5A | 1531 | 1.549 | 0.2289 | Yes | ||
| 23 | LPHN3 | 1559 | 1.538 | 0.2361 | Yes | ||
| 24 | KLF7 | 1567 | 1.535 | 0.2442 | Yes | ||
| 25 | BMF | 1578 | 1.530 | 0.2521 | Yes | ||
| 26 | POU4F2 | 1618 | 1.512 | 0.2586 | Yes | ||
| 27 | DMTF1 | 1633 | 1.505 | 0.2662 | Yes | ||
| 28 | CSNK1A1 | 1654 | 1.498 | 0.2735 | Yes | ||
| 29 | ZIC1 | 1684 | 1.489 | 0.2803 | Yes | ||
| 30 | RNF12 | 1689 | 1.488 | 0.2883 | Yes | ||
| 31 | PB1 | 2211 | 1.292 | 0.2715 | Yes | ||
| 32 | HOXB3 | 2354 | 1.240 | 0.2718 | Yes | ||
| 33 | SOX5 | 2456 | 1.209 | 0.2737 | Yes | ||
| 34 | GTF2A1 | 2485 | 1.202 | 0.2790 | Yes | ||
| 35 | ACACA | 2581 | 1.180 | 0.2812 | Yes | ||
| 36 | NRXN3 | 2622 | 1.170 | 0.2857 | Yes | ||
| 37 | ACVR2B | 2632 | 1.168 | 0.2917 | Yes | ||
| 38 | ID4 | 2702 | 1.151 | 0.2949 | Yes | ||
| 39 | EYA1 | 2734 | 1.143 | 0.2997 | Yes | ||
| 40 | MAP4K5 | 3156 | 1.048 | 0.2861 | No | ||
| 41 | NFIB | 3359 | 0.999 | 0.2823 | No | ||
| 42 | IGF1 | 3560 | 0.957 | 0.2784 | No | ||
| 43 | BCL11A | 3568 | 0.955 | 0.2833 | No | ||
| 44 | EPS8 | 3882 | 0.894 | 0.2738 | No | ||
| 45 | GRIK3 | 4007 | 0.871 | 0.2729 | No | ||
| 46 | STAG2 | 4119 | 0.852 | 0.2725 | No | ||
| 47 | HIPK1 | 4689 | 0.757 | 0.2505 | No | ||
| 48 | KIAA1128 | 4771 | 0.745 | 0.2509 | No | ||
| 49 | IL1RAPL1 | 4833 | 0.734 | 0.2521 | No | ||
| 50 | IRS4 | 5194 | 0.681 | 0.2394 | No | ||
| 51 | RHEBL1 | 5436 | 0.644 | 0.2318 | No | ||
| 52 | FGF12 | 5464 | 0.640 | 0.2341 | No | ||
| 53 | PDCD4 | 5639 | 0.616 | 0.2295 | No | ||
| 54 | PBEF1 | 5684 | 0.610 | 0.2308 | No | ||
| 55 | HOXB4 | 5849 | 0.581 | 0.2265 | No | ||
| 56 | PAPPA | 5897 | 0.575 | 0.2275 | No | ||
| 57 | OLFML3 | 6058 | 0.551 | 0.2231 | No | ||
| 58 | MAF1 | 6137 | 0.538 | 0.2225 | No | ||
| 59 | IGSF4B | 6201 | 0.526 | 0.2225 | No | ||
| 60 | SLC7A3 | 6435 | 0.495 | 0.2145 | No | ||
| 61 | CBFB | 6656 | 0.465 | 0.2070 | No | ||
| 62 | SRGAP2 | 6928 | 0.429 | 0.1969 | No | ||
| 63 | NEUROG2 | 6994 | 0.421 | 0.1962 | No | ||
| 64 | HOXB6 | 7009 | 0.419 | 0.1979 | No | ||
| 65 | PITX2 | 7310 | 0.378 | 0.1862 | No | ||
| 66 | EFNA1 | 8068 | 0.278 | 0.1530 | No | ||
| 67 | HEY1 | 8736 | 0.185 | 0.1234 | No | ||
| 68 | TNFRSF8 | 9043 | 0.140 | 0.1101 | No | ||
| 69 | WNT9A | 9222 | 0.117 | 0.1026 | No | ||
| 70 | PHEX | 9298 | 0.105 | 0.0997 | No | ||
| 71 | GPR50 | 9425 | 0.085 | 0.0944 | No | ||
| 72 | FBXO11 | 9480 | 0.077 | 0.0923 | No | ||
| 73 | DYRK1B | 9653 | 0.051 | 0.0847 | No | ||
| 74 | SLC5A3 | 10125 | -0.023 | 0.0632 | No | ||
| 75 | FGF6 | 10261 | -0.044 | 0.0573 | No | ||
| 76 | BLNK | 10309 | -0.050 | 0.0554 | No | ||
| 77 | MLLT7 | 10380 | -0.062 | 0.0525 | No | ||
| 78 | FOXA2 | 10563 | -0.088 | 0.0447 | No | ||
| 79 | HOXB8 | 10913 | -0.143 | 0.0294 | No | ||
| 80 | DNAJB7 | 11138 | -0.173 | 0.0201 | No | ||
| 81 | CCNG2 | 11327 | -0.197 | 0.0125 | No | ||
| 82 | CACNA1D | 11368 | -0.202 | 0.0118 | No | ||
| 83 | TBXAS1 | 11377 | -0.203 | 0.0126 | No | ||
| 84 | FILIP1 | 11515 | -0.219 | 0.0075 | No | ||
| 85 | LHX6 | 11544 | -0.222 | 0.0074 | No | ||
| 86 | TBX5 | 11638 | -0.232 | 0.0044 | No | ||
| 87 | OTP | 11653 | -0.234 | 0.0051 | No | ||
| 88 | NAV3 | 11788 | -0.248 | 0.0003 | No | ||
| 89 | MEOX1 | 12069 | -0.280 | -0.0111 | No | ||
| 90 | TREX2 | 12193 | -0.294 | -0.0151 | No | ||
| 91 | CACNA1G | 12380 | -0.318 | -0.0219 | No | ||
| 92 | EMCN | 12436 | -0.324 | -0.0226 | No | ||
| 93 | KLF15 | 12605 | -0.345 | -0.0284 | No | ||
| 94 | ACADSB | 12741 | -0.360 | -0.0327 | No | ||
| 95 | NEUROD2 | 13050 | -0.397 | -0.0446 | No | ||
| 96 | TMEFF1 | 13429 | -0.443 | -0.0595 | No | ||
| 97 | NKX2-2 | 13482 | -0.449 | -0.0595 | No | ||
| 98 | GNG11 | 13521 | -0.453 | -0.0587 | No | ||
| 99 | PPM2C | 13595 | -0.460 | -0.0596 | No | ||
| 100 | HNRPA0 | 14248 | -0.539 | -0.0865 | No | ||
| 101 | LMO3 | 15146 | -0.645 | -0.1241 | No | ||
| 102 | APC | 15491 | -0.686 | -0.1362 | No | ||
| 103 | PRKAG1 | 15526 | -0.689 | -0.1340 | No | ||
| 104 | AMELX | 15607 | -0.702 | -0.1338 | No | ||
| 105 | ITGA3 | 15609 | -0.702 | -0.1300 | No | ||
| 106 | GABARAP | 15640 | -0.707 | -0.1275 | No | ||
| 107 | OAZ2 | 15727 | -0.719 | -0.1275 | No | ||
| 108 | OTX2 | 16124 | -0.774 | -0.1414 | No | ||
| 109 | EFNB1 | 16280 | -0.795 | -0.1442 | No | ||
| 110 | PNMA1 | 16302 | -0.798 | -0.1408 | No | ||
| 111 | BARHL1 | 16398 | -0.810 | -0.1407 | No | ||
| 112 | DNALI1 | 16630 | -0.839 | -0.1467 | No | ||
| 113 | HAS2 | 16701 | -0.847 | -0.1453 | No | ||
| 114 | NR3C2 | 16760 | -0.856 | -0.1432 | No | ||
| 115 | KCNC2 | 16858 | -0.869 | -0.1429 | No | ||
| 116 | REPS2 | 17170 | -0.918 | -0.1522 | No | ||
| 117 | PIK3R3 | 17183 | -0.920 | -0.1477 | No | ||
| 118 | SEMA3A | 17659 | -1.000 | -0.1640 | No | ||
| 119 | CBX4 | 17710 | -1.008 | -0.1607 | No | ||
| 120 | SALL3 | 17753 | -1.014 | -0.1571 | No | ||
| 121 | ITGB8 | 17931 | -1.048 | -0.1595 | No | ||
| 122 | SMARCA2 | 17995 | -1.057 | -0.1566 | No | ||
| 123 | LRCH2 | 18034 | -1.064 | -0.1525 | No | ||
| 124 | POLG2 | 18066 | -1.070 | -0.1481 | No | ||
| 125 | DEDD | 18165 | -1.086 | -0.1466 | No | ||
| 126 | NTRK2 | 18173 | -1.087 | -0.1410 | No | ||
| 127 | ROBO1 | 18781 | -1.214 | -0.1622 | No | ||
| 128 | NFIX | 18888 | -1.239 | -0.1603 | No | ||
| 129 | HDAC9 | 19004 | -1.263 | -0.1586 | No | ||
| 130 | WASF2 | 19052 | -1.271 | -0.1538 | No | ||
| 131 | NRP2 | 19124 | -1.288 | -0.1500 | No | ||
| 132 | SRPX | 19132 | -1.291 | -0.1432 | No | ||
| 133 | RBBP6 | 19244 | -1.322 | -0.1411 | No | ||
| 134 | GDNF | 19313 | -1.344 | -0.1368 | No | ||
| 135 | EYA4 | 19386 | -1.362 | -0.1327 | No | ||
| 136 | SFRS6 | 19426 | -1.372 | -0.1270 | No | ||
| 137 | CALM3 | 19591 | -1.421 | -0.1267 | No | ||
| 138 | NR2F1 | 19622 | -1.432 | -0.1202 | No | ||
| 139 | ZIC4 | 20073 | -1.599 | -0.1321 | No | ||
| 140 | POU4F3 | 20078 | -1.600 | -0.1235 | No | ||
| 141 | MSRA | 20279 | -1.688 | -0.1234 | No | ||
| 142 | MNT | 20472 | -1.778 | -0.1225 | No | ||
| 143 | SGK | 20592 | -1.843 | -0.1179 | No | ||
| 144 | RFX1 | 20626 | -1.862 | -0.1092 | No | ||
| 145 | MTMR3 | 20790 | -1.964 | -0.1059 | No | ||
| 146 | NR2F2 | 21012 | -2.123 | -0.1044 | No | ||
| 147 | PRKCBP1 | 21031 | -2.135 | -0.0935 | No | ||
| 148 | GBF1 | 21377 | -2.528 | -0.0955 | No | ||
| 149 | JUNB | 21440 | -2.670 | -0.0837 | No | ||
| 150 | TAPBP | 21470 | -2.730 | -0.0701 | No | ||
| 151 | MAP4K4 | 21744 | -3.794 | -0.0618 | No | ||
| 152 | RNF122 | 21761 | -3.923 | -0.0411 | No | ||
| 153 | PIM2 | 21799 | -4.257 | -0.0194 | No | ||
| 154 | SALL2 | 21852 | -4.777 | 0.0044 | No |