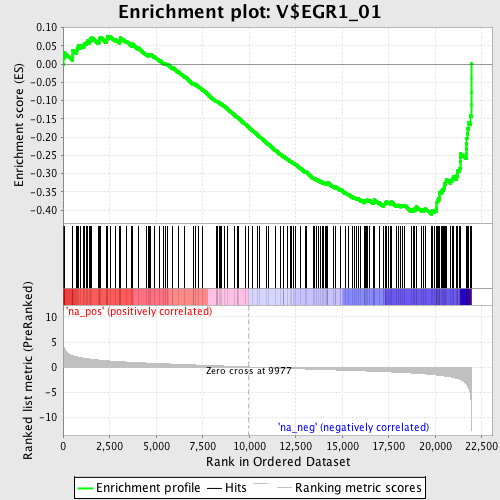
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$EGR1_01 |
| Enrichment Score (ES) | -0.4122698 |
| Normalized Enrichment Score (NES) | -1.8409151 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.010061766 |
| FWER p-Value | 0.075 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ORC4L | 29 | 4.329 | 0.0166 | No | ||
| 2 | MATR3 | 55 | 3.775 | 0.0311 | No | ||
| 3 | PAFAH1B1 | 513 | 2.259 | 0.0195 | No | ||
| 4 | RUNX1 | 523 | 2.244 | 0.0283 | No | ||
| 5 | NLK | 525 | 2.243 | 0.0376 | No | ||
| 6 | SFRS10 | 698 | 2.057 | 0.0382 | No | ||
| 7 | XPR1 | 752 | 2.008 | 0.0441 | No | ||
| 8 | USP1 | 805 | 1.961 | 0.0498 | No | ||
| 9 | TAF11 | 957 | 1.849 | 0.0506 | No | ||
| 10 | CAMK2A | 1103 | 1.758 | 0.0512 | No | ||
| 11 | ATE1 | 1146 | 1.734 | 0.0564 | No | ||
| 12 | TCTA | 1239 | 1.688 | 0.0592 | No | ||
| 13 | HMGA1 | 1301 | 1.652 | 0.0633 | No | ||
| 14 | GPHN | 1432 | 1.593 | 0.0639 | No | ||
| 15 | SPRED1 | 1466 | 1.574 | 0.0689 | No | ||
| 16 | YWHAE | 1550 | 1.540 | 0.0715 | No | ||
| 17 | FUS | 1896 | 1.396 | 0.0614 | No | ||
| 18 | PBX1 | 1941 | 1.379 | 0.0651 | No | ||
| 19 | DDX6 | 1948 | 1.376 | 0.0705 | No | ||
| 20 | SDCCAG8 | 1998 | 1.361 | 0.0739 | No | ||
| 21 | UCHL3 | 2352 | 1.240 | 0.0628 | No | ||
| 22 | NUMBL | 2356 | 1.240 | 0.0678 | No | ||
| 23 | CLTC | 2387 | 1.228 | 0.0715 | No | ||
| 24 | SH3KBP1 | 2392 | 1.226 | 0.0764 | No | ||
| 25 | PCTK2 | 2547 | 1.187 | 0.0743 | No | ||
| 26 | TBX3 | 2805 | 1.126 | 0.0671 | No | ||
| 27 | RNF24 | 3021 | 1.077 | 0.0617 | No | ||
| 28 | FBXL11 | 3036 | 1.074 | 0.0655 | No | ||
| 29 | SOCS5 | 3096 | 1.061 | 0.0672 | No | ||
| 30 | KLF3 | 3097 | 1.061 | 0.0716 | No | ||
| 31 | RPS6KA3 | 3383 | 0.995 | 0.0626 | No | ||
| 32 | ETV4 | 3689 | 0.931 | 0.0525 | No | ||
| 33 | RALGPS2 | 3719 | 0.926 | 0.0550 | No | ||
| 34 | KLHL10 | 4034 | 0.866 | 0.0441 | No | ||
| 35 | CACNA1E | 4498 | 0.787 | 0.0261 | No | ||
| 36 | CGGBP1 | 4612 | 0.770 | 0.0241 | No | ||
| 37 | VEGF | 4635 | 0.767 | 0.0263 | No | ||
| 38 | HNRPDL | 4715 | 0.753 | 0.0258 | No | ||
| 39 | CNOT6L | 4899 | 0.726 | 0.0204 | No | ||
| 40 | MYB | 5181 | 0.683 | 0.0103 | No | ||
| 41 | VGF | 5406 | 0.648 | 0.0027 | No | ||
| 42 | EIF5A | 5477 | 0.638 | 0.0021 | No | ||
| 43 | MSI1 | 5630 | 0.618 | -0.0023 | No | ||
| 44 | CTCF | 5879 | 0.578 | -0.0113 | No | ||
| 45 | SEP15 | 5887 | 0.576 | -0.0092 | No | ||
| 46 | ICAM5 | 6182 | 0.531 | -0.0205 | No | ||
| 47 | IMPDH1 | 6547 | 0.481 | -0.0353 | No | ||
| 48 | VTN | 7002 | 0.420 | -0.0544 | No | ||
| 49 | SSTR3 | 7025 | 0.417 | -0.0537 | No | ||
| 50 | HHEX | 7127 | 0.403 | -0.0566 | No | ||
| 51 | KCNS2 | 7288 | 0.381 | -0.0624 | No | ||
| 52 | UBXD3 | 7515 | 0.351 | -0.0713 | No | ||
| 53 | SP1 | 8214 | 0.260 | -0.1023 | No | ||
| 54 | HAS1 | 8251 | 0.254 | -0.1029 | No | ||
| 55 | RAB26 | 8277 | 0.250 | -0.1031 | No | ||
| 56 | ADSS | 8299 | 0.247 | -0.1030 | No | ||
| 57 | GNB2 | 8378 | 0.236 | -0.1056 | No | ||
| 58 | KCNN2 | 8469 | 0.222 | -0.1088 | No | ||
| 59 | CNNM4 | 8492 | 0.218 | -0.1089 | No | ||
| 60 | KCNQ5 | 8660 | 0.195 | -0.1158 | No | ||
| 61 | ATP1B2 | 8845 | 0.169 | -0.1235 | No | ||
| 62 | SMARCA5 | 9226 | 0.117 | -0.1405 | No | ||
| 63 | FAF1 | 9349 | 0.098 | -0.1457 | No | ||
| 64 | ZNHIT1 | 9389 | 0.090 | -0.1471 | No | ||
| 65 | FEV | 9433 | 0.084 | -0.1488 | No | ||
| 66 | SH3GL3 | 9799 | 0.027 | -0.1654 | No | ||
| 67 | RAB39 | 9957 | 0.003 | -0.1726 | No | ||
| 68 | GSH1 | 10176 | -0.030 | -0.1825 | No | ||
| 69 | AP3S1 | 10442 | -0.069 | -0.1944 | No | ||
| 70 | LRFN5 | 10538 | -0.084 | -0.1984 | No | ||
| 71 | HOXB8 | 10913 | -0.143 | -0.2150 | No | ||
| 72 | HCFC1R1 | 11022 | -0.159 | -0.2193 | No | ||
| 73 | EGLN2 | 11397 | -0.205 | -0.2356 | No | ||
| 74 | FZD1 | 11662 | -0.235 | -0.2468 | No | ||
| 75 | PORCN | 11839 | -0.256 | -0.2538 | No | ||
| 76 | HMGN2 | 11848 | -0.257 | -0.2531 | No | ||
| 77 | NFYC | 11857 | -0.258 | -0.2524 | No | ||
| 78 | HYAL2 | 12082 | -0.281 | -0.2616 | No | ||
| 79 | DLX4 | 12229 | -0.298 | -0.2670 | No | ||
| 80 | MRC2 | 12259 | -0.302 | -0.2671 | No | ||
| 81 | PRPF3 | 12389 | -0.319 | -0.2717 | No | ||
| 82 | GLRA3 | 12485 | -0.329 | -0.2747 | No | ||
| 83 | KCNAB3 | 12752 | -0.362 | -0.2854 | No | ||
| 84 | KCNB1 | 13032 | -0.395 | -0.2966 | No | ||
| 85 | NEUROD2 | 13050 | -0.397 | -0.2958 | No | ||
| 86 | EPHB1 | 13469 | -0.447 | -0.3131 | No | ||
| 87 | CDH2 | 13529 | -0.455 | -0.3139 | No | ||
| 88 | GPR132 | 13635 | -0.466 | -0.3168 | No | ||
| 89 | ACE | 13747 | -0.479 | -0.3199 | No | ||
| 90 | PDCD8 | 13851 | -0.493 | -0.3226 | No | ||
| 91 | HOXA2 | 13955 | -0.504 | -0.3253 | No | ||
| 92 | HOXA7 | 14011 | -0.510 | -0.3257 | No | ||
| 93 | CREB3L1 | 14108 | -0.520 | -0.3280 | No | ||
| 94 | ZBTB5 | 14161 | -0.528 | -0.3282 | No | ||
| 95 | TNFRSF12A | 14180 | -0.530 | -0.3268 | No | ||
| 96 | HMGN1 | 14227 | -0.535 | -0.3267 | No | ||
| 97 | KCNH5 | 14229 | -0.536 | -0.3245 | No | ||
| 98 | RFX4 | 14543 | -0.575 | -0.3365 | No | ||
| 99 | CDKN2C | 14633 | -0.587 | -0.3382 | No | ||
| 100 | MORF4L2 | 14644 | -0.589 | -0.3362 | No | ||
| 101 | SLC22A17 | 14877 | -0.615 | -0.3443 | No | ||
| 102 | SLC25A23 | 14926 | -0.621 | -0.3439 | No | ||
| 103 | CSMD3 | 15169 | -0.649 | -0.3524 | No | ||
| 104 | SREBF2 | 15318 | -0.668 | -0.3564 | No | ||
| 105 | PRKAG1 | 15526 | -0.689 | -0.3630 | No | ||
| 106 | PCOLCE | 15630 | -0.706 | -0.3648 | No | ||
| 107 | DLL4 | 15750 | -0.721 | -0.3673 | No | ||
| 108 | DDX5 | 15849 | -0.734 | -0.3688 | No | ||
| 109 | PLOD3 | 15996 | -0.757 | -0.3724 | No | ||
| 110 | OTX1 | 16181 | -0.782 | -0.3776 | No | ||
| 111 | AKAP12 | 16182 | -0.782 | -0.3743 | No | ||
| 112 | DVL2 | 16252 | -0.790 | -0.3742 | No | ||
| 113 | CLPTM1 | 16308 | -0.798 | -0.3734 | No | ||
| 114 | TPBG | 16360 | -0.805 | -0.3725 | No | ||
| 115 | ARL1 | 16438 | -0.815 | -0.3726 | No | ||
| 116 | BRD2 | 16659 | -0.843 | -0.3792 | No | ||
| 117 | NRK | 16693 | -0.847 | -0.3772 | No | ||
| 118 | EFEMP2 | 16720 | -0.851 | -0.3749 | No | ||
| 119 | CBLN1 | 16735 | -0.853 | -0.3720 | No | ||
| 120 | PPARGC1B | 16989 | -0.889 | -0.3800 | No | ||
| 121 | GLTSCR2 | 17234 | -0.928 | -0.3873 | No | ||
| 122 | HSPB9 | 17240 | -0.929 | -0.3837 | No | ||
| 123 | SOX12 | 17297 | -0.937 | -0.3824 | No | ||
| 124 | HRK | 17305 | -0.939 | -0.3788 | No | ||
| 125 | RHOB | 17370 | -0.948 | -0.3778 | No | ||
| 126 | RHOA | 17480 | -0.969 | -0.3788 | No | ||
| 127 | GCN5L2 | 17601 | -0.990 | -0.3802 | No | ||
| 128 | PRKACA | 17631 | -0.996 | -0.3774 | No | ||
| 129 | ITGB8 | 17931 | -1.048 | -0.3868 | No | ||
| 130 | HOXA5 | 18015 | -1.061 | -0.3863 | No | ||
| 131 | PDGFB | 18137 | -1.080 | -0.3873 | No | ||
| 132 | SMYD5 | 18225 | -1.098 | -0.3868 | No | ||
| 133 | LYPLA2 | 18325 | -1.120 | -0.3867 | No | ||
| 134 | SERTAD1 | 18702 | -1.195 | -0.3990 | No | ||
| 135 | CACNA1A | 18814 | -1.221 | -0.3991 | No | ||
| 136 | GRB2 | 18879 | -1.237 | -0.3969 | No | ||
| 137 | TPM4 | 18962 | -1.254 | -0.3954 | No | ||
| 138 | GIT1 | 18987 | -1.260 | -0.3913 | No | ||
| 139 | RBBP6 | 19244 | -1.322 | -0.3976 | No | ||
| 140 | HR | 19382 | -1.361 | -0.3983 | No | ||
| 141 | APLP2 | 19476 | -1.388 | -0.3968 | No | ||
| 142 | APP | 19814 | -1.503 | -0.4060 | Yes | ||
| 143 | PCDH17 | 19838 | -1.509 | -0.4008 | Yes | ||
| 144 | VAMP2 | 19969 | -1.554 | -0.4004 | Yes | ||
| 145 | AP1G1 | 20050 | -1.590 | -0.3975 | Yes | ||
| 146 | KCND2 | 20081 | -1.602 | -0.3922 | Yes | ||
| 147 | PCSK2 | 20085 | -1.604 | -0.3857 | Yes | ||
| 148 | SEC14L1 | 20086 | -1.604 | -0.3790 | Yes | ||
| 149 | FOXP2 | 20114 | -1.616 | -0.3736 | Yes | ||
| 150 | CMAS | 20156 | -1.632 | -0.3687 | Yes | ||
| 151 | GRIN1 | 20215 | -1.658 | -0.3645 | Yes | ||
| 152 | MEF2C | 20238 | -1.670 | -0.3586 | Yes | ||
| 153 | ERF | 20240 | -1.671 | -0.3517 | Yes | ||
| 154 | CLSTN1 | 20310 | -1.704 | -0.3478 | Yes | ||
| 155 | SCN8A | 20403 | -1.742 | -0.3448 | Yes | ||
| 156 | SF1 | 20461 | -1.773 | -0.3401 | Yes | ||
| 157 | MNT | 20472 | -1.778 | -0.3332 | Yes | ||
| 158 | MYST2 | 20483 | -1.782 | -0.3263 | Yes | ||
| 159 | ELL | 20572 | -1.835 | -0.3227 | Yes | ||
| 160 | TLE3 | 20574 | -1.836 | -0.3152 | Yes | ||
| 161 | BAHD1 | 20822 | -1.978 | -0.3183 | Yes | ||
| 162 | KCNC1 | 20897 | -2.031 | -0.3133 | Yes | ||
| 163 | KLF16 | 20992 | -2.112 | -0.3089 | Yes | ||
| 164 | SH3BP1 | 21143 | -2.229 | -0.3065 | Yes | ||
| 165 | MAN2A2 | 21198 | -2.293 | -0.2995 | Yes | ||
| 166 | UBTF | 21212 | -2.303 | -0.2905 | Yes | ||
| 167 | DNAJB5 | 21315 | -2.438 | -0.2851 | Yes | ||
| 168 | MAP3K4 | 21340 | -2.470 | -0.2760 | Yes | ||
| 169 | RASGEF1A | 21341 | -2.470 | -0.2658 | Yes | ||
| 170 | ARF3 | 21349 | -2.483 | -0.2558 | Yes | ||
| 171 | ZFPM1 | 21370 | -2.515 | -0.2463 | Yes | ||
| 172 | MGLL | 21658 | -3.302 | -0.2458 | Yes | ||
| 173 | NRGN | 21664 | -3.322 | -0.2323 | Yes | ||
| 174 | TRIB1 | 21684 | -3.431 | -0.2189 | Yes | ||
| 175 | TLE4 | 21695 | -3.506 | -0.2048 | Yes | ||
| 176 | CORO1C | 21703 | -3.548 | -0.1905 | Yes | ||
| 177 | RELB | 21749 | -3.826 | -0.1767 | Yes | ||
| 178 | CNN2 | 21797 | -4.241 | -0.1613 | Yes | ||
| 179 | PJA1 | 21889 | -5.620 | -0.1422 | Yes | ||
| 180 | NAB2 | 21936 | -8.017 | -0.1111 | Yes | ||
| 181 | LEF1 | 21939 | -8.298 | -0.0768 | Yes | ||
| 182 | EGR1 | 21942 | -8.847 | -0.0402 | Yes | ||
| 183 | EGR3 | 21943 | -9.744 | 0.0002 | Yes |