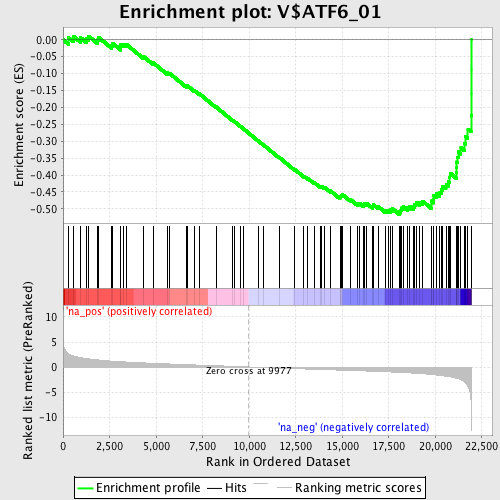
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$ATF6_01 |
| Enrichment Score (ES) | -0.5165904 |
| Normalized Enrichment Score (NES) | -2.1040685 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.378685E-4 |
| FWER p-Value | 0.0010 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GFRA1 | 292 | 2.629 | 0.0060 | No | ||
| 2 | ELF1 | 551 | 2.205 | 0.0103 | No | ||
| 3 | ATBF1 | 949 | 1.852 | 0.0058 | No | ||
| 4 | STAG1 | 1261 | 1.679 | 0.0039 | No | ||
| 5 | TNKS2 | 1370 | 1.618 | 0.0108 | No | ||
| 6 | GRIA3 | 1846 | 1.415 | -0.0005 | No | ||
| 7 | TLOC1 | 1901 | 1.391 | 0.0072 | No | ||
| 8 | PAX1 | 2611 | 1.173 | -0.0166 | No | ||
| 9 | RORC | 2672 | 1.158 | -0.0108 | No | ||
| 10 | TOP1 | 3099 | 1.061 | -0.0225 | No | ||
| 11 | SRP19 | 3103 | 1.060 | -0.0149 | No | ||
| 12 | SYNCRIP | 3238 | 1.028 | -0.0135 | No | ||
| 13 | LHX5 | 3409 | 0.989 | -0.0140 | No | ||
| 14 | NOL4 | 4345 | 0.814 | -0.0508 | No | ||
| 15 | IL1RAPL1 | 4833 | 0.734 | -0.0677 | No | ||
| 16 | FLNC | 5586 | 0.624 | -0.0975 | No | ||
| 17 | SLC39A5 | 5722 | 0.603 | -0.0992 | No | ||
| 18 | PTOV1 | 6626 | 0.470 | -0.1371 | No | ||
| 19 | TRPC1 | 6683 | 0.461 | -0.1363 | No | ||
| 20 | DHX40 | 7061 | 0.411 | -0.1505 | No | ||
| 21 | PITX2 | 7310 | 0.378 | -0.1591 | No | ||
| 22 | SP1 | 8214 | 0.260 | -0.1985 | No | ||
| 23 | HOXB9 | 9119 | 0.131 | -0.2389 | No | ||
| 24 | TWIST1 | 9198 | 0.120 | -0.2416 | No | ||
| 25 | IRX3 | 9548 | 0.068 | -0.2570 | No | ||
| 26 | SEC23A | 9672 | 0.047 | -0.2623 | No | ||
| 27 | SLC30A5 | 10473 | -0.075 | -0.2984 | No | ||
| 28 | LMX1B | 10774 | -0.120 | -0.3112 | No | ||
| 29 | ADAMTS3 | 11609 | -0.229 | -0.3477 | No | ||
| 30 | SLC39A13 | 12455 | -0.326 | -0.3839 | No | ||
| 31 | LHX1 | 12937 | -0.383 | -0.4031 | No | ||
| 32 | SFRS1 | 13144 | -0.407 | -0.4096 | No | ||
| 33 | NKX2-2 | 13482 | -0.449 | -0.4217 | No | ||
| 34 | TPT1 | 13830 | -0.490 | -0.4339 | No | ||
| 35 | MRPS18B | 13900 | -0.498 | -0.4334 | No | ||
| 36 | PPP1R10 | 14070 | -0.516 | -0.4374 | No | ||
| 37 | KDELR1 | 14368 | -0.553 | -0.4469 | No | ||
| 38 | SLC22A17 | 14877 | -0.615 | -0.4656 | No | ||
| 39 | KDELR3 | 14927 | -0.621 | -0.4633 | No | ||
| 40 | KLF13 | 14944 | -0.623 | -0.4595 | No | ||
| 41 | GRIN2B | 15010 | -0.629 | -0.4578 | No | ||
| 42 | NANS | 15426 | -0.680 | -0.4718 | No | ||
| 43 | HCRTR2 | 15824 | -0.732 | -0.4846 | No | ||
| 44 | SCYL1 | 15912 | -0.743 | -0.4831 | No | ||
| 45 | TRIM39 | 16151 | -0.777 | -0.4883 | No | ||
| 46 | RAB2 | 16207 | -0.785 | -0.4851 | No | ||
| 47 | OSBP | 16315 | -0.799 | -0.4841 | No | ||
| 48 | GSC | 16615 | -0.837 | -0.4916 | No | ||
| 49 | BRD2 | 16659 | -0.843 | -0.4874 | No | ||
| 50 | C1QTNF7 | 16942 | -0.883 | -0.4938 | No | ||
| 51 | XPNPEP1 | 17306 | -0.939 | -0.5035 | No | ||
| 52 | LENG9 | 17465 | -0.966 | -0.5037 | No | ||
| 53 | ARF4 | 17597 | -0.989 | -0.5024 | No | ||
| 54 | USP2 | 17696 | -1.007 | -0.4995 | No | ||
| 55 | SH2D2A | 18071 | -1.071 | -0.5087 | Yes | ||
| 56 | NPTX1 | 18150 | -1.084 | -0.5043 | Yes | ||
| 57 | NTRK2 | 18173 | -1.087 | -0.4974 | Yes | ||
| 58 | GOLGB1 | 18280 | -1.109 | -0.4941 | Yes | ||
| 59 | EIF4A1 | 18503 | -1.154 | -0.4957 | Yes | ||
| 60 | KDELR2 | 18615 | -1.178 | -0.4922 | Yes | ||
| 61 | PHC1 | 18842 | -1.226 | -0.4935 | Yes | ||
| 62 | GRB2 | 18879 | -1.237 | -0.4861 | Yes | ||
| 63 | TPM4 | 18962 | -1.254 | -0.4806 | Yes | ||
| 64 | KCNA6 | 19164 | -1.300 | -0.4802 | Yes | ||
| 65 | GDNF | 19313 | -1.344 | -0.4771 | Yes | ||
| 66 | ICA1 | 19782 | -1.491 | -0.4876 | Yes | ||
| 67 | DLST | 19783 | -1.492 | -0.4767 | Yes | ||
| 68 | SEMA3B | 19915 | -1.535 | -0.4714 | Yes | ||
| 69 | PER1 | 19921 | -1.537 | -0.4603 | Yes | ||
| 70 | GNG4 | 20044 | -1.587 | -0.4542 | Yes | ||
| 71 | ERF | 20240 | -1.671 | -0.4509 | Yes | ||
| 72 | TYRO3 | 20318 | -1.705 | -0.4419 | Yes | ||
| 73 | TBX20 | 20399 | -1.741 | -0.4328 | Yes | ||
| 74 | TLE3 | 20574 | -1.836 | -0.4272 | Yes | ||
| 75 | CREM | 20689 | -1.903 | -0.4185 | Yes | ||
| 76 | SYT11 | 20779 | -1.957 | -0.4082 | Yes | ||
| 77 | SEC24D | 20788 | -1.963 | -0.3941 | Yes | ||
| 78 | LMO4 | 21122 | -2.212 | -0.3931 | Yes | ||
| 79 | STAT3 | 21136 | -2.226 | -0.3774 | Yes | ||
| 80 | ARMCX2 | 21145 | -2.230 | -0.3614 | Yes | ||
| 81 | MADD | 21207 | -2.298 | -0.3473 | Yes | ||
| 82 | ETV6 | 21235 | -2.325 | -0.3314 | Yes | ||
| 83 | GBF1 | 21377 | -2.528 | -0.3193 | Yes | ||
| 84 | GCC1 | 21552 | -2.901 | -0.3060 | Yes | ||
| 85 | RAB6IP1 | 21609 | -3.116 | -0.2856 | Yes | ||
| 86 | DUSP4 | 21754 | -3.844 | -0.2640 | Yes | ||
| 87 | NR4A1 | 21923 | -6.633 | -0.2230 | Yes | ||
| 88 | EGR1 | 21942 | -8.847 | -0.1588 | Yes | ||
| 89 | EGR3 | 21943 | -9.744 | -0.0873 | Yes | ||
| 90 | EGR2 | 21944 | -11.902 | 0.0001 | Yes |
