Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$AR_Q6 |
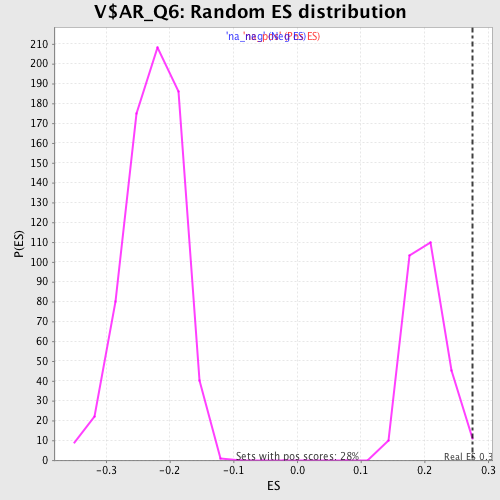
| Enrichment Score (ES) | 0.27504796 |
| Normalized Enrichment Score (NES) | 1.3612579 |
| Nominal p-value | 0.017921148 |
| FDR q-value | 0.20264465 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL2RA | 15 | 4.747 | 0.0243 | Yes | ||
| 2 | SFRS12 | 33 | 4.169 | 0.0456 | Yes | ||
| 3 | EPHA2 | 46 | 3.838 | 0.0652 | Yes | ||
| 4 | AP2B1 | 273 | 2.682 | 0.0690 | Yes | ||
| 5 | CORO2A | 343 | 2.525 | 0.0792 | Yes | ||
| 6 | H2AFZ | 345 | 2.524 | 0.0924 | Yes | ||
| 7 | PUM2 | 472 | 2.322 | 0.0989 | Yes | ||
| 8 | TBL1X | 480 | 2.303 | 0.1107 | Yes | ||
| 9 | ORC1L | 505 | 2.267 | 0.1216 | Yes | ||
| 10 | SH3BGRL | 683 | 2.077 | 0.1244 | Yes | ||
| 11 | PURA | 828 | 1.932 | 0.1280 | Yes | ||
| 12 | TLK1 | 843 | 1.923 | 0.1375 | Yes | ||
| 13 | CASK | 873 | 1.906 | 0.1462 | Yes | ||
| 14 | DNAJC7 | 1087 | 1.766 | 0.1457 | Yes | ||
| 15 | PCF11 | 1156 | 1.729 | 0.1517 | Yes | ||
| 16 | SLC13A1 | 1183 | 1.715 | 0.1596 | Yes | ||
| 17 | MBNL1 | 1213 | 1.702 | 0.1672 | Yes | ||
| 18 | ANGPT1 | 1254 | 1.681 | 0.1743 | Yes | ||
| 19 | MAP2K5 | 1264 | 1.672 | 0.1827 | Yes | ||
| 20 | FST | 1383 | 1.611 | 0.1858 | Yes | ||
| 21 | TIAL1 | 1439 | 1.589 | 0.1916 | Yes | ||
| 22 | SKP2 | 1455 | 1.579 | 0.1992 | Yes | ||
| 23 | SLC25A25 | 1514 | 1.556 | 0.2048 | Yes | ||
| 24 | TOM1L2 | 1601 | 1.521 | 0.2089 | Yes | ||
| 25 | TM9SF2 | 1610 | 1.516 | 0.2165 | Yes | ||
| 26 | ARHGAP26 | 1690 | 1.488 | 0.2207 | Yes | ||
| 27 | PPARGC1A | 1723 | 1.472 | 0.2270 | Yes | ||
| 28 | TFDP2 | 1796 | 1.440 | 0.2313 | Yes | ||
| 29 | MST1R | 1824 | 1.425 | 0.2376 | Yes | ||
| 30 | GRIA3 | 1846 | 1.415 | 0.2441 | Yes | ||
| 31 | WDFY3 | 2082 | 1.334 | 0.2403 | Yes | ||
| 32 | MTF1 | 2118 | 1.323 | 0.2457 | Yes | ||
| 33 | PB1 | 2211 | 1.292 | 0.2483 | Yes | ||
| 34 | BCL2L1 | 2231 | 1.283 | 0.2542 | Yes | ||
| 35 | USP54 | 2414 | 1.221 | 0.2523 | Yes | ||
| 36 | ATRNL1 | 2478 | 1.204 | 0.2557 | Yes | ||
| 37 | GAP43 | 2480 | 1.203 | 0.2620 | Yes | ||
| 38 | HTR4 | 2568 | 1.183 | 0.2643 | Yes | ||
| 39 | BNC2 | 2586 | 1.180 | 0.2697 | Yes | ||
| 40 | NRXN3 | 2622 | 1.170 | 0.2743 | Yes | ||
| 41 | SCN3B | 2743 | 1.140 | 0.2748 | Yes | ||
| 42 | DYRK2 | 3002 | 1.082 | 0.2686 | Yes | ||
| 43 | SLC12A1 | 3093 | 1.062 | 0.2701 | Yes | ||
| 44 | SYNCRIP | 3238 | 1.028 | 0.2689 | Yes | ||
| 45 | SNX2 | 3392 | 0.992 | 0.2671 | Yes | ||
| 46 | LHX5 | 3409 | 0.989 | 0.2716 | Yes | ||
| 47 | PRM1 | 3629 | 0.943 | 0.2665 | Yes | ||
| 48 | FA2H | 3655 | 0.938 | 0.2703 | Yes | ||
| 49 | SUPT16H | 3661 | 0.936 | 0.2750 | Yes | ||
| 50 | ANGPTL2 | 3881 | 0.894 | 0.2697 | No | ||
| 51 | SLC30A2 | 3919 | 0.886 | 0.2727 | No | ||
| 52 | KLHL10 | 4034 | 0.866 | 0.2720 | No | ||
| 53 | RAPGEF6 | 4129 | 0.850 | 0.2722 | No | ||
| 54 | NR6A1 | 4514 | 0.784 | 0.2587 | No | ||
| 55 | UBE2D3 | 4979 | 0.711 | 0.2412 | No | ||
| 56 | CX36 | 5107 | 0.695 | 0.2390 | No | ||
| 57 | LMO1 | 5118 | 0.693 | 0.2422 | No | ||
| 58 | UGCGL1 | 5566 | 0.626 | 0.2250 | No | ||
| 59 | ID1 | 6328 | 0.509 | 0.1927 | No | ||
| 60 | EPN1 | 6822 | 0.442 | 0.1724 | No | ||
| 61 | HOXD3 | 6935 | 0.428 | 0.1695 | No | ||
| 62 | DHX40 | 7061 | 0.411 | 0.1660 | No | ||
| 63 | PDYN | 7191 | 0.395 | 0.1621 | No | ||
| 64 | PITX2 | 7310 | 0.378 | 0.1587 | No | ||
| 65 | BHLHB5 | 7453 | 0.360 | 0.1541 | No | ||
| 66 | SOX14 | 7656 | 0.331 | 0.1466 | No | ||
| 67 | LMOD1 | 7657 | 0.331 | 0.1483 | No | ||
| 68 | MTRF1L | 7740 | 0.322 | 0.1462 | No | ||
| 69 | PLA2G2F | 7962 | 0.293 | 0.1376 | No | ||
| 70 | TEK | 8113 | 0.271 | 0.1322 | No | ||
| 71 | UFC1 | 8295 | 0.248 | 0.1252 | No | ||
| 72 | TNNI1 | 8554 | 0.211 | 0.1145 | No | ||
| 73 | TGM5 | 8563 | 0.209 | 0.1152 | No | ||
| 74 | QTRTD1 | 8667 | 0.194 | 0.1115 | No | ||
| 75 | RPL28 | 8794 | 0.176 | 0.1066 | No | ||
| 76 | SOS2 | 9624 | 0.055 | 0.0689 | No | ||
| 77 | BTK | 9783 | 0.031 | 0.0618 | No | ||
| 78 | GPRC6A | 9910 | 0.011 | 0.0560 | No | ||
| 79 | VPS45A | 10138 | -0.024 | 0.0458 | No | ||
| 80 | RPA3 | 10199 | -0.033 | 0.0432 | No | ||
| 81 | KRTAP8-1 | 10221 | -0.036 | 0.0424 | No | ||
| 82 | SGK2 | 11096 | -0.167 | 0.0032 | No | ||
| 83 | BMP7 | 11682 | -0.237 | -0.0224 | No | ||
| 84 | ACTR3 | 11907 | -0.263 | -0.0313 | No | ||
| 85 | SLC22A6 | 12289 | -0.308 | -0.0472 | No | ||
| 86 | YAP1 | 12825 | -0.370 | -0.0698 | No | ||
| 87 | RHEB | 13014 | -0.393 | -0.0764 | No | ||
| 88 | NKIRAS2 | 13128 | -0.405 | -0.0794 | No | ||
| 89 | SMOC1 | 13129 | -0.405 | -0.0773 | No | ||
| 90 | SHH | 13258 | -0.419 | -0.0809 | No | ||
| 91 | NKX2-2 | 13482 | -0.449 | -0.0888 | No | ||
| 92 | POU2F3 | 13591 | -0.459 | -0.0913 | No | ||
| 93 | ANKRD2 | 13766 | -0.482 | -0.0968 | No | ||
| 94 | HOXA2 | 13955 | -0.504 | -0.1028 | No | ||
| 95 | GDAP1L1 | 13958 | -0.504 | -0.1002 | No | ||
| 96 | HOXC6 | 14117 | -0.521 | -0.1047 | No | ||
| 97 | SEZ6 | 14220 | -0.535 | -0.1066 | No | ||
| 98 | VAX1 | 14334 | -0.549 | -0.1088 | No | ||
| 99 | PPP2R2B | 14403 | -0.557 | -0.1090 | No | ||
| 100 | PPP1R3D | 14533 | -0.574 | -0.1119 | No | ||
| 101 | SLC22A17 | 14877 | -0.615 | -0.1244 | No | ||
| 102 | KCNJ2 | 14969 | -0.625 | -0.1253 | No | ||
| 103 | HOXA3 | 15093 | -0.638 | -0.1276 | No | ||
| 104 | CITED1 | 15150 | -0.646 | -0.1268 | No | ||
| 105 | CSMD3 | 15169 | -0.649 | -0.1242 | No | ||
| 106 | NBEA | 15279 | -0.662 | -0.1257 | No | ||
| 107 | MAPK3 | 15401 | -0.677 | -0.1277 | No | ||
| 108 | H2AFJ | 15414 | -0.679 | -0.1246 | No | ||
| 109 | PRKAG1 | 15526 | -0.689 | -0.1261 | No | ||
| 110 | MYLK | 15649 | -0.708 | -0.1279 | No | ||
| 111 | DLL4 | 15750 | -0.721 | -0.1287 | No | ||
| 112 | LIF | 16250 | -0.790 | -0.1475 | No | ||
| 113 | PCDH9 | 16469 | -0.819 | -0.1532 | No | ||
| 114 | CD34 | 16470 | -0.819 | -0.1488 | No | ||
| 115 | NEO1 | 16777 | -0.858 | -0.1584 | No | ||
| 116 | SPATA2 | 17166 | -0.918 | -0.1713 | No | ||
| 117 | NDST2 | 17285 | -0.935 | -0.1718 | No | ||
| 118 | AP1GBP1 | 17288 | -0.935 | -0.1670 | No | ||
| 119 | LDB2 | 17543 | -0.980 | -0.1735 | No | ||
| 120 | IHPK2 | 17628 | -0.995 | -0.1721 | No | ||
| 121 | SLC9A9 | 17716 | -1.009 | -0.1707 | No | ||
| 122 | MID1 | 17719 | -1.010 | -0.1655 | No | ||
| 123 | LLGL2 | 17842 | -1.029 | -0.1657 | No | ||
| 124 | EPHA7 | 17870 | -1.036 | -0.1615 | No | ||
| 125 | ITGB8 | 17931 | -1.048 | -0.1587 | No | ||
| 126 | PHOX2B | 18114 | -1.078 | -0.1614 | No | ||
| 127 | FGA | 18228 | -1.098 | -0.1607 | No | ||
| 128 | LUZP1 | 18276 | -1.109 | -0.1571 | No | ||
| 129 | SMAD1 | 18330 | -1.120 | -0.1536 | No | ||
| 130 | SRPK1 | 18472 | -1.148 | -0.1540 | No | ||
| 131 | A2BP1 | 18581 | -1.170 | -0.1528 | No | ||
| 132 | DDX17 | 18608 | -1.175 | -0.1478 | No | ||
| 133 | HDAC9 | 19004 | -1.263 | -0.1593 | No | ||
| 134 | FBS1 | 19095 | -1.281 | -0.1566 | No | ||
| 135 | SESN3 | 19220 | -1.317 | -0.1554 | No | ||
| 136 | RBBP6 | 19244 | -1.322 | -0.1495 | No | ||
| 137 | NR2F1 | 19622 | -1.432 | -0.1592 | No | ||
| 138 | ARMET | 19642 | -1.437 | -0.1525 | No | ||
| 139 | KCNMB1 | 19719 | -1.468 | -0.1483 | No | ||
| 140 | GSR | 19832 | -1.507 | -0.1454 | No | ||
| 141 | LRRK1 | 19845 | -1.511 | -0.1380 | No | ||
| 142 | SLC22A2 | 19871 | -1.518 | -0.1312 | No | ||
| 143 | ELAVL2 | 20020 | -1.577 | -0.1296 | No | ||
| 144 | CRY1 | 20063 | -1.594 | -0.1232 | No | ||
| 145 | FOXP2 | 20114 | -1.616 | -0.1169 | No | ||
| 146 | TPM3 | 20175 | -1.642 | -0.1110 | No | ||
| 147 | MEF2C | 20238 | -1.670 | -0.1051 | No | ||
| 148 | PYGO2 | 20296 | -1.696 | -0.0987 | No | ||
| 149 | DMD | 20507 | -1.799 | -0.0989 | No | ||
| 150 | SLC25A12 | 20633 | -1.866 | -0.0948 | No | ||
| 151 | NNAT | 20797 | -1.968 | -0.0919 | No | ||
| 152 | ZFYVE26 | 20817 | -1.975 | -0.0823 | No | ||
| 153 | SIN3A | 20857 | -2.001 | -0.0736 | No | ||
| 154 | LRRN5 | 20888 | -2.026 | -0.0643 | No | ||
| 155 | HSD17B2 | 20932 | -2.058 | -0.0554 | No | ||
| 156 | MAPKAPK5 | 20969 | -2.083 | -0.0461 | No | ||
| 157 | JUP | 21149 | -2.234 | -0.0425 | No | ||
| 158 | CAMSAP1 | 21407 | -2.583 | -0.0407 | No | ||
| 159 | TEGT | 21474 | -2.738 | -0.0293 | No | ||
| 160 | SH2D3C | 21529 | -2.856 | -0.0167 | No | ||
| 161 | ELMO1 | 21628 | -3.167 | -0.0045 | No | ||
| 162 | PTGER2 | 21715 | -3.615 | 0.0106 | No |