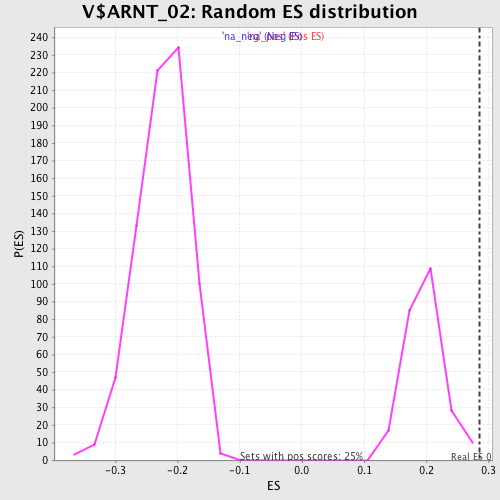
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$ARNT_02 |
| Enrichment Score (ES) | 0.28552404 |
| Normalized Enrichment Score (NES) | 1.4410282 |
| Nominal p-value | 0.004016064 |
| FDR q-value | 0.18794483 |
| FWER p-Value | 0.904 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TRPM7 | 70 | 3.654 | 0.0153 | Yes | ||
| 2 | AGRP | 133 | 3.235 | 0.0288 | Yes | ||
| 3 | IGF2R | 156 | 3.130 | 0.0436 | Yes | ||
| 4 | SNX16 | 174 | 2.997 | 0.0579 | Yes | ||
| 5 | RABGAP1 | 196 | 2.942 | 0.0719 | Yes | ||
| 6 | CBX5 | 300 | 2.613 | 0.0803 | Yes | ||
| 7 | ANKRD17 | 307 | 2.602 | 0.0932 | Yes | ||
| 8 | H2AFZ | 345 | 2.524 | 0.1043 | Yes | ||
| 9 | ANKHD1 | 395 | 2.431 | 0.1143 | Yes | ||
| 10 | SLC43A1 | 420 | 2.395 | 0.1253 | Yes | ||
| 11 | ATF7IP | 496 | 2.279 | 0.1334 | Yes | ||
| 12 | ATP7A | 569 | 2.185 | 0.1411 | Yes | ||
| 13 | TNRC15 | 668 | 2.088 | 0.1472 | Yes | ||
| 14 | EPC1 | 736 | 2.022 | 0.1543 | Yes | ||
| 15 | HNRPA1 | 802 | 1.963 | 0.1613 | Yes | ||
| 16 | PURA | 828 | 1.932 | 0.1699 | Yes | ||
| 17 | ATP6V1H | 860 | 1.911 | 0.1781 | Yes | ||
| 18 | GNAS | 956 | 1.850 | 0.1831 | Yes | ||
| 19 | AATF | 1086 | 1.766 | 0.1861 | Yes | ||
| 20 | RBBP4 | 1209 | 1.704 | 0.1891 | Yes | ||
| 21 | PICALM | 1210 | 1.703 | 0.1977 | Yes | ||
| 22 | UBE2B | 1228 | 1.693 | 0.2055 | Yes | ||
| 23 | HIRA | 1291 | 1.658 | 0.2110 | Yes | ||
| 24 | HMGA1 | 1301 | 1.652 | 0.2190 | Yes | ||
| 25 | STX6 | 1339 | 1.636 | 0.2256 | Yes | ||
| 26 | GTF2H1 | 1438 | 1.590 | 0.2291 | Yes | ||
| 27 | CD164 | 1470 | 1.573 | 0.2356 | Yes | ||
| 28 | DIRC2 | 1474 | 1.572 | 0.2434 | Yes | ||
| 29 | NOTCH1 | 1500 | 1.562 | 0.2502 | Yes | ||
| 30 | ZIC1 | 1684 | 1.489 | 0.2493 | Yes | ||
| 31 | CANX | 1790 | 1.442 | 0.2518 | Yes | ||
| 32 | ARMCX3 | 1884 | 1.401 | 0.2546 | Yes | ||
| 33 | HIF1A | 1894 | 1.396 | 0.2612 | Yes | ||
| 34 | PDCD6IP | 2096 | 1.330 | 0.2587 | Yes | ||
| 35 | SOCS2 | 2303 | 1.259 | 0.2556 | Yes | ||
| 36 | CLTC | 2387 | 1.228 | 0.2580 | Yes | ||
| 37 | SLC35A5 | 2598 | 1.177 | 0.2543 | Yes | ||
| 38 | PRKCE | 2610 | 1.173 | 0.2597 | Yes | ||
| 39 | NCL | 2655 | 1.163 | 0.2636 | Yes | ||
| 40 | VLDLR | 2659 | 1.162 | 0.2693 | Yes | ||
| 41 | RNF146 | 2696 | 1.153 | 0.2735 | Yes | ||
| 42 | SIRT1 | 2780 | 1.133 | 0.2754 | Yes | ||
| 43 | CPEB4 | 2861 | 1.114 | 0.2774 | Yes | ||
| 44 | CPT1A | 2903 | 1.106 | 0.2811 | Yes | ||
| 45 | CUL5 | 2977 | 1.089 | 0.2832 | Yes | ||
| 46 | CAMK2D | 3046 | 1.072 | 0.2855 | Yes | ||
| 47 | SYNCRIP | 3238 | 1.028 | 0.2819 | No | ||
| 48 | TOPORS | 3384 | 0.994 | 0.2803 | No | ||
| 49 | SNX2 | 3392 | 0.992 | 0.2850 | No | ||
| 50 | ZMYND12 | 3748 | 0.918 | 0.2733 | No | ||
| 51 | NDUFA7 | 3811 | 0.907 | 0.2751 | No | ||
| 52 | SNTB2 | 4014 | 0.870 | 0.2702 | No | ||
| 53 | USP31 | 4397 | 0.804 | 0.2567 | No | ||
| 54 | PTPRF | 4405 | 0.802 | 0.2604 | No | ||
| 55 | KIAA1715 | 4684 | 0.759 | 0.2515 | No | ||
| 56 | HPS3 | 4686 | 0.758 | 0.2553 | No | ||
| 57 | CNNM1 | 5026 | 0.704 | 0.2433 | No | ||
| 58 | IRS4 | 5194 | 0.681 | 0.2391 | No | ||
| 59 | GMFB | 5373 | 0.653 | 0.2342 | No | ||
| 60 | VGF | 5406 | 0.648 | 0.2360 | No | ||
| 61 | SLCO1C1 | 5515 | 0.633 | 0.2342 | No | ||
| 62 | NFATC3 | 5945 | 0.567 | 0.2174 | No | ||
| 63 | FEN1 | 5963 | 0.565 | 0.2194 | No | ||
| 64 | RAB24 | 6083 | 0.548 | 0.2168 | No | ||
| 65 | ICAM5 | 6182 | 0.531 | 0.2149 | No | ||
| 66 | BAX | 6537 | 0.482 | 0.2011 | No | ||
| 67 | EN1 | 6629 | 0.469 | 0.1993 | No | ||
| 68 | RGL1 | 6759 | 0.451 | 0.1956 | No | ||
| 69 | HOXB5 | 7102 | 0.406 | 0.1820 | No | ||
| 70 | RCOR2 | 7245 | 0.388 | 0.1774 | No | ||
| 71 | HRH3 | 7342 | 0.373 | 0.1749 | No | ||
| 72 | EME1 | 7358 | 0.371 | 0.1761 | No | ||
| 73 | KCNE4 | 7744 | 0.321 | 0.1600 | No | ||
| 74 | GGN | 8022 | 0.285 | 0.1487 | No | ||
| 75 | U2AF2 | 8138 | 0.268 | 0.1448 | No | ||
| 76 | ATP6V1A | 8194 | 0.261 | 0.1436 | No | ||
| 77 | ASNA1 | 8211 | 0.260 | 0.1442 | No | ||
| 78 | GNB2 | 8378 | 0.236 | 0.1378 | No | ||
| 79 | LZTS2 | 8483 | 0.219 | 0.1341 | No | ||
| 80 | STMN1 | 8643 | 0.198 | 0.1278 | No | ||
| 81 | RPS2 | 9123 | 0.130 | 0.1064 | No | ||
| 82 | SLC38A5 | 9361 | 0.095 | 0.0960 | No | ||
| 83 | HNRPH2 | 9848 | 0.019 | 0.0738 | No | ||
| 84 | FOXD3 | 10024 | -0.009 | 0.0658 | No | ||
| 85 | IFRD2 | 10127 | -0.023 | 0.0612 | No | ||
| 86 | ALDH6A1 | 10147 | -0.026 | 0.0605 | No | ||
| 87 | TCEAL1 | 10350 | -0.058 | 0.0515 | No | ||
| 88 | MRPL27 | 10446 | -0.071 | 0.0475 | No | ||
| 89 | TRIM3 | 10679 | -0.104 | 0.0374 | No | ||
| 90 | RPS28 | 10788 | -0.122 | 0.0330 | No | ||
| 91 | FXYD6 | 11109 | -0.169 | 0.0192 | No | ||
| 92 | EGLN2 | 11397 | -0.205 | 0.0070 | No | ||
| 93 | ADAMTS3 | 11609 | -0.229 | -0.0015 | No | ||
| 94 | NR1D1 | 11759 | -0.246 | -0.0071 | No | ||
| 95 | REV1L | 11791 | -0.249 | -0.0073 | No | ||
| 96 | CLN3 | 11867 | -0.258 | -0.0094 | No | ||
| 97 | MAFF | 12071 | -0.280 | -0.0173 | No | ||
| 98 | RAD9A | 12139 | -0.288 | -0.0190 | No | ||
| 99 | ARPC5 | 12171 | -0.292 | -0.0189 | No | ||
| 100 | BLOC1S1 | 12177 | -0.292 | -0.0177 | No | ||
| 101 | ALG1 | 12301 | -0.309 | -0.0217 | No | ||
| 102 | SLCO5A1 | 12383 | -0.318 | -0.0239 | No | ||
| 103 | CENTB5 | 12387 | -0.319 | -0.0224 | No | ||
| 104 | APEX1 | 12641 | -0.349 | -0.0322 | No | ||
| 105 | FBXL19 | 12676 | -0.352 | -0.0320 | No | ||
| 106 | HPS5 | 12734 | -0.360 | -0.0328 | No | ||
| 107 | RRAGC | 12976 | -0.387 | -0.0419 | No | ||
| 108 | NEUROD2 | 13050 | -0.397 | -0.0433 | No | ||
| 109 | SMNDC1 | 13483 | -0.449 | -0.0609 | No | ||
| 110 | PIAS4 | 13555 | -0.456 | -0.0618 | No | ||
| 111 | CHRM1 | 13669 | -0.470 | -0.0646 | No | ||
| 112 | MRPL40 | 13752 | -0.480 | -0.0660 | No | ||
| 113 | ATP6V1C1 | 13844 | -0.492 | -0.0677 | No | ||
| 114 | PRSS15 | 13894 | -0.498 | -0.0674 | No | ||
| 115 | HOXA7 | 14011 | -0.510 | -0.0702 | No | ||
| 116 | ZBTB10 | 14106 | -0.520 | -0.0718 | No | ||
| 117 | SLC36A1 | 14534 | -0.574 | -0.0886 | No | ||
| 118 | RFX4 | 14543 | -0.575 | -0.0860 | No | ||
| 119 | CDKN2C | 14633 | -0.587 | -0.0871 | No | ||
| 120 | HOXA1 | 14710 | -0.597 | -0.0876 | No | ||
| 121 | RAB31 | 14807 | -0.607 | -0.0889 | No | ||
| 122 | ANAPC13 | 14862 | -0.614 | -0.0883 | No | ||
| 123 | OLFM2 | 15035 | -0.633 | -0.0930 | No | ||
| 124 | SEC23IP | 15108 | -0.640 | -0.0931 | No | ||
| 125 | BHLHB3 | 15178 | -0.650 | -0.0930 | No | ||
| 126 | OSGEP | 15221 | -0.654 | -0.0916 | No | ||
| 127 | MTCH2 | 15278 | -0.662 | -0.0908 | No | ||
| 128 | GATA4 | 15522 | -0.689 | -0.0985 | No | ||
| 129 | EIF4B | 15548 | -0.693 | -0.0962 | No | ||
| 130 | SPG21 | 15571 | -0.697 | -0.0937 | No | ||
| 131 | GPM6B | 15593 | -0.700 | -0.0911 | No | ||
| 132 | RAB30 | 15939 | -0.747 | -0.1032 | No | ||
| 133 | KBTBD2 | 16017 | -0.760 | -0.1028 | No | ||
| 134 | PRPS2 | 16194 | -0.783 | -0.1070 | No | ||
| 135 | LTBR | 16514 | -0.825 | -0.1175 | No | ||
| 136 | AVPI1 | 16525 | -0.826 | -0.1137 | No | ||
| 137 | HOXD10 | 16566 | -0.831 | -0.1114 | No | ||
| 138 | LAMP1 | 16617 | -0.837 | -0.1094 | No | ||
| 139 | BRD2 | 16659 | -0.843 | -0.1071 | No | ||
| 140 | CBX6 | 16868 | -0.871 | -0.1122 | No | ||
| 141 | HOXC5 | 16896 | -0.874 | -0.1090 | No | ||
| 142 | DSCAM | 16951 | -0.884 | -0.1071 | No | ||
| 143 | THUMPD2 | 17041 | -0.899 | -0.1066 | No | ||
| 144 | DLX2 | 17254 | -0.930 | -0.1116 | No | ||
| 145 | PA2G4 | 17267 | -0.932 | -0.1075 | No | ||
| 146 | HSPBAP1 | 17652 | -0.999 | -0.1201 | No | ||
| 147 | FABP3 | 17801 | -1.023 | -0.1217 | No | ||
| 148 | ELAVL3 | 18006 | -1.060 | -0.1257 | No | ||
| 149 | COMMD8 | 18012 | -1.061 | -0.1206 | No | ||
| 150 | WHSC1L1 | 18053 | -1.068 | -0.1170 | No | ||
| 151 | ATF4 | 18125 | -1.079 | -0.1148 | No | ||
| 152 | NPTX1 | 18150 | -1.084 | -0.1104 | No | ||
| 153 | CBARA1 | 18425 | -1.140 | -0.1173 | No | ||
| 154 | AKAP1 | 18464 | -1.147 | -0.1132 | No | ||
| 155 | NGFRAP1 | 18588 | -1.171 | -0.1129 | No | ||
| 156 | HOXA9 | 18656 | -1.186 | -0.1100 | No | ||
| 157 | EIF4G1 | 18692 | -1.193 | -0.1056 | No | ||
| 158 | HTATIP | 18792 | -1.215 | -0.1040 | No | ||
| 159 | PRDM4 | 18894 | -1.239 | -0.1024 | No | ||
| 160 | CDK5R1 | 19121 | -1.288 | -0.1063 | No | ||
| 161 | ARMET | 19642 | -1.437 | -0.1229 | No | ||
| 162 | RNF44 | 19686 | -1.455 | -0.1175 | No | ||
| 163 | PES1 | 19934 | -1.542 | -0.1210 | No | ||
| 164 | DCTN4 | 19985 | -1.562 | -0.1154 | No | ||
| 165 | UBE4B | 20303 | -1.700 | -0.1214 | No | ||
| 166 | HEXA | 20478 | -1.781 | -0.1204 | No | ||
| 167 | PSME3 | 20480 | -1.781 | -0.1114 | No | ||
| 168 | SGK | 20592 | -1.843 | -0.1072 | No | ||
| 169 | CSRP2BP | 20614 | -1.854 | -0.0988 | No | ||
| 170 | ZFYVE26 | 20817 | -1.975 | -0.0981 | No | ||
| 171 | GLA | 20870 | -2.009 | -0.0903 | No | ||
| 172 | TUBA1 | 20994 | -2.113 | -0.0853 | No | ||
| 173 | POLR3C | 21085 | -2.181 | -0.0784 | No | ||
| 174 | TGIF2 | 21166 | -2.254 | -0.0707 | No | ||
| 175 | NFX1 | 21251 | -2.345 | -0.0627 | No | ||
| 176 | INSM1 | 21275 | -2.374 | -0.0518 | No | ||
| 177 | TUBA4 | 21490 | -2.758 | -0.0476 | No | ||
| 178 | BDNF | 21491 | -2.758 | -0.0337 | No | ||
| 179 | SNN | 21655 | -3.277 | -0.0246 | No | ||
| 180 | CSK | 21693 | -3.473 | -0.0088 | No | ||
| 181 | BEX1 | 21774 | -4.033 | 0.0079 | No |