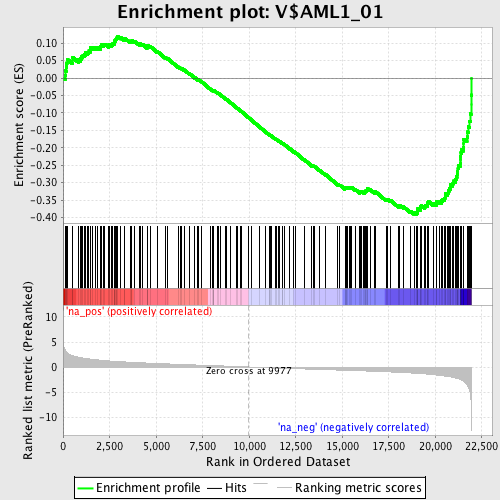
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$AML1_01 |
| Enrichment Score (ES) | -0.39120695 |
| Normalized Enrichment Score (NES) | -1.7621163 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.015313114 |
| FWER p-Value | 0.211 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TCF12 | 109 | 3.364 | 0.0089 | No | ||
| 2 | LCK | 137 | 3.203 | 0.0210 | No | ||
| 3 | PACS1 | 163 | 3.058 | 0.0325 | No | ||
| 4 | MMP14 | 192 | 2.944 | 0.0434 | No | ||
| 5 | IGSF4 | 212 | 2.865 | 0.0544 | No | ||
| 6 | ATF7IP | 496 | 2.279 | 0.0509 | No | ||
| 7 | RAG1 | 498 | 2.276 | 0.0603 | No | ||
| 8 | ATP2A2 | 836 | 1.927 | 0.0528 | No | ||
| 9 | PTK9 | 947 | 1.853 | 0.0554 | No | ||
| 10 | JMJD1C | 973 | 1.836 | 0.0619 | No | ||
| 11 | ARHGAP12 | 1064 | 1.777 | 0.0651 | No | ||
| 12 | PCF11 | 1156 | 1.729 | 0.0681 | No | ||
| 13 | PICALM | 1210 | 1.703 | 0.0727 | No | ||
| 14 | SCN2B | 1320 | 1.645 | 0.0745 | No | ||
| 15 | CYP17A1 | 1389 | 1.608 | 0.0781 | No | ||
| 16 | GZMB | 1463 | 1.577 | 0.0813 | No | ||
| 17 | CHES1 | 1467 | 1.573 | 0.0876 | No | ||
| 18 | BMF | 1578 | 1.530 | 0.0889 | No | ||
| 19 | FLI1 | 1719 | 1.474 | 0.0886 | No | ||
| 20 | RGS1 | 1848 | 1.415 | 0.0886 | No | ||
| 21 | MPL | 2009 | 1.359 | 0.0869 | No | ||
| 22 | MYOD1 | 2022 | 1.354 | 0.0919 | No | ||
| 23 | ANK3 | 2067 | 1.340 | 0.0955 | No | ||
| 24 | CBL | 2171 | 1.305 | 0.0961 | No | ||
| 25 | SLITRK5 | 2249 | 1.277 | 0.0979 | No | ||
| 26 | SOX5 | 2456 | 1.209 | 0.0934 | No | ||
| 27 | GTF2A1 | 2485 | 1.202 | 0.0971 | No | ||
| 28 | NRXN3 | 2622 | 1.170 | 0.0957 | No | ||
| 29 | RORC | 2672 | 1.158 | 0.0983 | No | ||
| 30 | SCN3B | 2743 | 1.140 | 0.0998 | No | ||
| 31 | AGTRL1 | 2776 | 1.133 | 0.1030 | No | ||
| 32 | SIRT1 | 2780 | 1.133 | 0.1076 | No | ||
| 33 | POU2F1 | 2833 | 1.121 | 0.1099 | No | ||
| 34 | ABCD4 | 2841 | 1.120 | 0.1142 | No | ||
| 35 | HOXC4 | 2893 | 1.107 | 0.1164 | No | ||
| 36 | GPD1 | 2904 | 1.106 | 0.1205 | No | ||
| 37 | SLC12A6 | 3085 | 1.064 | 0.1167 | No | ||
| 38 | TLL2 | 3292 | 1.014 | 0.1114 | No | ||
| 39 | RICS | 3308 | 1.011 | 0.1149 | No | ||
| 40 | NGB | 3605 | 0.948 | 0.1053 | No | ||
| 41 | SUPT16H | 3661 | 0.936 | 0.1066 | No | ||
| 42 | EIF3S1 | 3699 | 0.929 | 0.1088 | No | ||
| 43 | AKAP3 | 3844 | 0.902 | 0.1059 | No | ||
| 44 | SLC37A2 | 4122 | 0.851 | 0.0967 | No | ||
| 45 | JDP2 | 4145 | 0.848 | 0.0992 | No | ||
| 46 | PSMA1 | 4270 | 0.827 | 0.0969 | No | ||
| 47 | NUCB2 | 4511 | 0.785 | 0.0892 | No | ||
| 48 | DNM3 | 4528 | 0.782 | 0.0917 | No | ||
| 49 | PDE3B | 4535 | 0.780 | 0.0946 | No | ||
| 50 | HIPK1 | 4689 | 0.757 | 0.0907 | No | ||
| 51 | ARHGAP21 | 5068 | 0.699 | 0.0763 | No | ||
| 52 | MTX1 | 5475 | 0.638 | 0.0603 | No | ||
| 53 | CTSK | 5600 | 0.622 | 0.0571 | No | ||
| 54 | DAB1 | 6194 | 0.528 | 0.0321 | No | ||
| 55 | KCNJ1 | 6306 | 0.511 | 0.0291 | No | ||
| 56 | CUGBP1 | 6374 | 0.503 | 0.0281 | No | ||
| 57 | COL4A1 | 6519 | 0.484 | 0.0235 | No | ||
| 58 | MR1 | 6788 | 0.447 | 0.0130 | No | ||
| 59 | SCEL | 7080 | 0.409 | 0.0014 | No | ||
| 60 | RCOR2 | 7245 | 0.388 | -0.0046 | No | ||
| 61 | LMO2 | 7292 | 0.381 | -0.0051 | No | ||
| 62 | COL9A2 | 7408 | 0.366 | -0.0089 | No | ||
| 63 | SCNN1A | 7921 | 0.297 | -0.0312 | No | ||
| 64 | DDX23 | 8048 | 0.281 | -0.0358 | No | ||
| 65 | ATP6V0B | 8091 | 0.274 | -0.0366 | No | ||
| 66 | PDZK1 | 8093 | 0.274 | -0.0355 | No | ||
| 67 | UBL3 | 8096 | 0.273 | -0.0344 | No | ||
| 68 | ANXA8 | 8279 | 0.250 | -0.0418 | No | ||
| 69 | ITGA10 | 8335 | 0.241 | -0.0433 | No | ||
| 70 | RASAL2 | 8459 | 0.223 | -0.0480 | No | ||
| 71 | IL23A | 8716 | 0.188 | -0.0590 | No | ||
| 72 | ANKRD1 | 8783 | 0.178 | -0.0613 | No | ||
| 73 | BLOC1S2 | 8986 | 0.150 | -0.0700 | No | ||
| 74 | THBS3 | 9303 | 0.104 | -0.0840 | No | ||
| 75 | MMP13 | 9366 | 0.094 | -0.0865 | No | ||
| 76 | DIABLO | 9521 | 0.071 | -0.0933 | No | ||
| 77 | FOXD2 | 9581 | 0.064 | -0.0957 | No | ||
| 78 | RAB39 | 9957 | 0.003 | -0.1130 | No | ||
| 79 | FCER1G | 10142 | -0.025 | -0.1213 | No | ||
| 80 | CLDN10 | 10535 | -0.084 | -0.1390 | No | ||
| 81 | TACSTD2 | 10857 | -0.132 | -0.1532 | No | ||
| 82 | MATN1 | 11105 | -0.168 | -0.1638 | No | ||
| 83 | TNFSF4 | 11111 | -0.169 | -0.1633 | No | ||
| 84 | KCTD4 | 11159 | -0.175 | -0.1648 | No | ||
| 85 | MKRN3 | 11211 | -0.183 | -0.1664 | No | ||
| 86 | WNT8B | 11417 | -0.206 | -0.1749 | No | ||
| 87 | SLC6A9 | 11471 | -0.213 | -0.1765 | No | ||
| 88 | NRAS | 11559 | -0.223 | -0.1796 | No | ||
| 89 | TBX5 | 11638 | -0.232 | -0.1822 | No | ||
| 90 | BTG4 | 11782 | -0.248 | -0.1877 | No | ||
| 91 | NAV3 | 11788 | -0.248 | -0.1869 | No | ||
| 92 | EMP1 | 11871 | -0.259 | -0.1896 | No | ||
| 93 | BLOC1S1 | 12177 | -0.292 | -0.2024 | No | ||
| 94 | PGF | 12393 | -0.319 | -0.2110 | No | ||
| 95 | MOAP1 | 12487 | -0.329 | -0.2139 | No | ||
| 96 | ZBTB8 | 12968 | -0.387 | -0.2343 | No | ||
| 97 | ADAMTS4 | 13367 | -0.435 | -0.2508 | No | ||
| 98 | PDZRN4 | 13432 | -0.443 | -0.2519 | No | ||
| 99 | MAP3K11 | 13487 | -0.449 | -0.2525 | No | ||
| 100 | MAMDC1 | 13788 | -0.485 | -0.2643 | No | ||
| 101 | HOXC6 | 14117 | -0.521 | -0.2772 | No | ||
| 102 | NHLRC2 | 14764 | -0.602 | -0.3044 | No | ||
| 103 | TBK1 | 14869 | -0.614 | -0.3066 | No | ||
| 104 | LMO3 | 15146 | -0.645 | -0.3166 | No | ||
| 105 | HDGF | 15167 | -0.649 | -0.3149 | No | ||
| 106 | SFRS4 | 15222 | -0.654 | -0.3146 | No | ||
| 107 | ACSL5 | 15243 | -0.656 | -0.3128 | No | ||
| 108 | IFNG | 15303 | -0.665 | -0.3128 | No | ||
| 109 | NDUFA9 | 15362 | -0.672 | -0.3127 | No | ||
| 110 | CRTAC1 | 15450 | -0.683 | -0.3138 | No | ||
| 111 | SLC37A4 | 15513 | -0.688 | -0.3138 | No | ||
| 112 | ADAMTS8 | 15703 | -0.715 | -0.3195 | No | ||
| 113 | RAB30 | 15939 | -0.747 | -0.3272 | No | ||
| 114 | EIF2B3 | 15957 | -0.750 | -0.3249 | No | ||
| 115 | STRC | 16058 | -0.765 | -0.3263 | No | ||
| 116 | PTPN22 | 16138 | -0.775 | -0.3267 | No | ||
| 117 | COL4A2 | 16220 | -0.787 | -0.3272 | No | ||
| 118 | PXN | 16227 | -0.787 | -0.3242 | No | ||
| 119 | RIN1 | 16270 | -0.793 | -0.3229 | No | ||
| 120 | MIZF | 16324 | -0.800 | -0.3220 | No | ||
| 121 | INPPL1 | 16331 | -0.801 | -0.3189 | No | ||
| 122 | RGS18 | 16332 | -0.801 | -0.3156 | No | ||
| 123 | VRK1 | 16492 | -0.822 | -0.3195 | No | ||
| 124 | RAB5B | 16753 | -0.855 | -0.3279 | No | ||
| 125 | PAX6 | 16780 | -0.859 | -0.3255 | No | ||
| 126 | RDH5 | 17361 | -0.947 | -0.3483 | No | ||
| 127 | BCAR3 | 17447 | -0.961 | -0.3482 | No | ||
| 128 | PLXNC1 | 17586 | -0.988 | -0.3504 | No | ||
| 129 | LY9 | 18037 | -1.064 | -0.3667 | No | ||
| 130 | DGKA | 18084 | -1.073 | -0.3644 | No | ||
| 131 | LUZP1 | 18276 | -1.109 | -0.3685 | No | ||
| 132 | DKK1 | 18669 | -1.188 | -0.3816 | No | ||
| 133 | GRASP | 18874 | -1.234 | -0.3859 | No | ||
| 134 | NOTCH2 | 18991 | -1.260 | -0.3860 | Yes | ||
| 135 | NFRKB | 19029 | -1.267 | -0.3824 | Yes | ||
| 136 | EIF2C1 | 19041 | -1.270 | -0.3777 | Yes | ||
| 137 | MRPS31 | 19062 | -1.274 | -0.3733 | Yes | ||
| 138 | PDE6H | 19189 | -1.306 | -0.3737 | Yes | ||
| 139 | ENTPD1 | 19196 | -1.308 | -0.3685 | Yes | ||
| 140 | RHOG | 19258 | -1.326 | -0.3658 | Yes | ||
| 141 | AP1G2 | 19423 | -1.372 | -0.3677 | Yes | ||
| 142 | XCL1 | 19473 | -1.388 | -0.3642 | Yes | ||
| 143 | ACIN1 | 19579 | -1.419 | -0.3631 | Yes | ||
| 144 | TITF1 | 19597 | -1.423 | -0.3580 | Yes | ||
| 145 | SNX1 | 19641 | -1.437 | -0.3540 | Yes | ||
| 146 | FGD4 | 19877 | -1.520 | -0.3585 | Yes | ||
| 147 | SLC15A3 | 20046 | -1.589 | -0.3596 | Yes | ||
| 148 | CRY1 | 20063 | -1.594 | -0.3538 | Yes | ||
| 149 | ARNT | 20204 | -1.654 | -0.3533 | Yes | ||
| 150 | ID3 | 20325 | -1.709 | -0.3518 | Yes | ||
| 151 | SCN8A | 20403 | -1.742 | -0.3481 | Yes | ||
| 152 | TPM1 | 20488 | -1.784 | -0.3446 | Yes | ||
| 153 | DACT1 | 20524 | -1.809 | -0.3387 | Yes | ||
| 154 | HLX1 | 20533 | -1.816 | -0.3315 | Yes | ||
| 155 | SEMA4A | 20669 | -1.891 | -0.3299 | Yes | ||
| 156 | CREM | 20689 | -1.903 | -0.3228 | Yes | ||
| 157 | FGF4 | 20738 | -1.930 | -0.3170 | Yes | ||
| 158 | CD69 | 20802 | -1.970 | -0.3118 | Yes | ||
| 159 | CAP1 | 20831 | -1.986 | -0.3048 | Yes | ||
| 160 | EDARADD | 20942 | -2.063 | -0.3013 | Yes | ||
| 161 | KIF1B | 20997 | -2.115 | -0.2950 | Yes | ||
| 162 | GPATC2 | 21084 | -2.179 | -0.2900 | Yes | ||
| 163 | DNAJC14 | 21135 | -2.225 | -0.2830 | Yes | ||
| 164 | TNNT2 | 21167 | -2.256 | -0.2751 | Yes | ||
| 165 | MADD | 21207 | -2.298 | -0.2674 | Yes | ||
| 166 | IBRDC3 | 21217 | -2.307 | -0.2582 | Yes | ||
| 167 | GRK5 | 21250 | -2.344 | -0.2500 | Yes | ||
| 168 | HIVEP3 | 21332 | -2.463 | -0.2435 | Yes | ||
| 169 | RASGEF1A | 21341 | -2.470 | -0.2336 | Yes | ||
| 170 | ARF3 | 21349 | -2.483 | -0.2236 | Yes | ||
| 171 | CD6 | 21376 | -2.526 | -0.2144 | Yes | ||
| 172 | PRICKLE1 | 21424 | -2.616 | -0.2057 | Yes | ||
| 173 | BATF | 21502 | -2.787 | -0.1976 | Yes | ||
| 174 | STAT2 | 21522 | -2.837 | -0.1868 | Yes | ||
| 175 | S100A9 | 21527 | -2.855 | -0.1751 | Yes | ||
| 176 | CORO1C | 21703 | -3.548 | -0.1684 | Yes | ||
| 177 | SLC2A3 | 21731 | -3.704 | -0.1543 | Yes | ||
| 178 | ARHGEF2 | 21769 | -3.994 | -0.1395 | Yes | ||
| 179 | ITGB7 | 21847 | -4.721 | -0.1234 | Yes | ||
| 180 | VIM | 21890 | -5.636 | -0.1020 | Yes | ||
| 181 | NR4A1 | 21923 | -6.633 | -0.0760 | Yes | ||
| 182 | TACC2 | 21924 | -6.663 | -0.0483 | Yes | ||
| 183 | EGR2 | 21944 | -11.902 | 0.0001 | Yes |